|
1
|
Im SS, Seo J, You JE, Bang HW, Kim Y, Kweon J, Kim Y, Shin DM, Son J. BIX01294 suppresses PDAC growth through inhibition of glutaminase-mediated glutathione dynamics. Mol Metab 2025; 94:102113. [PMID: 39961401 PMCID: PMC11905835 DOI: 10.1016/j.molmet.2025.102113] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Journal Information] [Submit a Manuscript] [Subscribe] [Scholar Register] [Received: 10/04/2024] [Revised: 02/05/2025] [Accepted: 02/11/2025] [Indexed: 02/23/2025] Open
Abstract
OBJECTIVES Increased expression of glutaminase (GLS) has been found to correlate with more aggressive disease and poorer prognosis in patients with several types of cancer, including breast, lung, and pancreatic cancer. G9a histone methyltransferase inhibitors may have anticancer activity. The present study assessed whether BIX01294 (BIX), a G9a histone methyltransferase inhibitor, can inhibit glutaminase (GLS) in pancreatic ductal adenocarcinoma (PDAC) cells. METHODS The effects of BIX on mitochondrial metabolism in PDAC cells were evaluated by targeted liquid chromatography-tandem mass spectrometry (LC-MS/MS) metabolomic analysis. To assess the impact of BIX on glutathione dynamics, real-time changes in glutathione levels were monitored by FreSHtracer-based GSH assays. RESULTS BIX significantly inhibited the growth of PDAC cells, both in vitro and in vivo, and robustly induced apoptotic cell death. BIX significantly increased the cellular NADP+/NADPH ratio and decreased the ratio of reduced-to-oxidized glutathione (GSH:GSSG). In addition, BIX decreased GSH levels and increased ROS levels. N-acetyl-l-cysteine (NAC) supplementation dramatically rescued PDAC cells from BIX-induced apoptosis. Furthermore, BIX inhibited the transcription of GLS by inhibiting Jumonji-domain histone demethylases but not G9a histone methyltransferase. One Jumonji-domain histone demethylase, KDM6B, epigenetically regulated GLS expression by binding to the GLS gene promoter. CONCLUSIONS Collectively, these findings suggest that BIX could be a potent therapeutic agent in patients with PDAC through its inhibition of GLS-mediated cellular redox balance.
Collapse
Affiliation(s)
- Se Seul Im
- Department of Biochemistry and Molecular Biology, Brain Korea 21 project, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, South Korea
| | - Jihyeon Seo
- Department of Biochemistry and Molecular Biology, Brain Korea 21 project, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, South Korea
| | - Ji Eun You
- Department of Biochemistry and Molecular Biology, Brain Korea 21 project, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, South Korea
| | - Hye Won Bang
- Department of Biochemistry and Molecular Biology, Brain Korea 21 project, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, South Korea
| | - YongHwan Kim
- Department of Cell and Genetic Engineering, Brain Korea 21 project, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, South Korea
| | - Jiyeon Kweon
- Department of Cell and Genetic Engineering, Brain Korea 21 project, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, South Korea
| | - Yongsub Kim
- Department of Cell and Genetic Engineering, Brain Korea 21 project, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, South Korea
| | - Dong-Myung Shin
- Department of Cell and Genetic Engineering, Brain Korea 21 project, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, South Korea
| | - Jaekyoung Son
- Department of Biochemistry and Molecular Biology, Brain Korea 21 project, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, South Korea.
| |
Collapse
|
|
2
|
Chiang JC, Shang Z, Rosales T, Cai L, Chen WM, Cai F, Vu H, Minna JD, Ni M, Davis AJ, Timmerman RD, DeBerardinis RJ, Zhang Y. Lipoylation inhibition enhances radiation control of lung cancer by suppressing homologous recombination DNA damage repair. SCIENCE ADVANCES 2025; 11:eadt1241. [PMID: 40073141 PMCID: PMC11900879 DOI: 10.1126/sciadv.adt1241] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [MESH Headings] [Grants] [Track Full Text] [Download PDF] [Figures] [Subscribe] [Scholar Register] [Received: 09/12/2024] [Accepted: 02/05/2025] [Indexed: 03/14/2025]
Abstract
Lung cancer exhibits altered metabolism, influencing its response to radiation. To investigate the metabolic regulation of radiation response, we conducted a comprehensive, metabolic-wide CRISPR-Cas9 loss-of-function screen using radiation as selection pressure in human non-small cell lung cancer. Lipoylation emerged as a key metabolic target for radiosensitization, with lipoyltransferase 1 (LIPT1) identified as a top hit. LIPT1 covalently conjugates mitochondrial 2-ketoacid dehydrogenases with lipoic acid, facilitating enzymatic functions involved in the tricarboxylic acid cycle. Inhibiting lipoylation, either through genetic LIPT1 knockout or a lipoylation inhibitor (CPI-613), enhanced tumor control by radiation. Mechanistically, lipoylation inhibition increased 2-hydroxyglutarate, leading to H3K9 trimethylation, disrupting TIP60 recruitment and ataxia telangiectasia mutated (ATM)-mediated DNA damage repair signaling, impairing homologous recombination repair. In summary, our findings reveal a critical role of LIPT1 in regulating DNA damage and chromosome stability and may suggest a means to enhance therapeutic outcomes with DNA-damaging agents.
Collapse
Affiliation(s)
- Jui-Chung Chiang
- Department of Radiation Oncology, Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA
| | - Zengfu Shang
- Department of Radiation Oncology, Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA
| | - Tracy Rosales
- Howard Hughes Medical Institute, Eugene McDermott Center for Human Growth and Development, and Children’s Medical Center Research Institute, University of Texas Southwestern Medical Center, Dallas, TX 75235, USA
| | - Ling Cai
- Peter O’Donnell, Jr. School of Public Health, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA
| | - Wei-Min Chen
- Department of Radiation Oncology, Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA
| | - Feng Cai
- Howard Hughes Medical Institute, Eugene McDermott Center for Human Growth and Development, and Children’s Medical Center Research Institute, University of Texas Southwestern Medical Center, Dallas, TX 75235, USA
| | - Hieu Vu
- Howard Hughes Medical Institute, Eugene McDermott Center for Human Growth and Development, and Children’s Medical Center Research Institute, University of Texas Southwestern Medical Center, Dallas, TX 75235, USA
| | - John D. Minna
- Hamon Center for Therapeutic Oncology Research, Departments of Internal Medicine and Pharmacology, Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA
| | - Min Ni
- Howard Hughes Medical Institute, Eugene McDermott Center for Human Growth and Development, and Children’s Medical Center Research Institute, University of Texas Southwestern Medical Center, Dallas, TX 75235, USA
| | - Anthony J. Davis
- Department of Radiation Oncology, Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA
| | - Robert D. Timmerman
- Department of Radiation Oncology, Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA
| | - Ralph J. DeBerardinis
- Howard Hughes Medical Institute, Eugene McDermott Center for Human Growth and Development, and Children’s Medical Center Research Institute, University of Texas Southwestern Medical Center, Dallas, TX 75235, USA
| | - Yuanyuan Zhang
- Department of Radiation Oncology, Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA
| |
Collapse
|
|
3
|
Kuras M, Betancourt LH, Hong R, Szadai L, Rodriguez J, Horvatovich P, Pla I, Eriksson J, Szeitz B, Deszcz B, Welinder C, Sugihara Y, Ekedahl H, Baldetorp B, Ingvar C, Lundgren L, Lindberg H, Oskolas H, Horvath Z, Rezeli M, Gil J, Appelqvist R, Kemény LV, Malm J, Sanchez A, Szasz AM, Pawłowski K, Wieslander E, Fenyö D, Nemeth IB, Marko-Varga G. Proteogenomic Profiling of Treatment-Naïve Metastatic Malignant Melanoma. Cancers (Basel) 2025; 17:832. [PMID: 40075679 PMCID: PMC11899103 DOI: 10.3390/cancers17050832] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Key Words] [Grants] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 01/24/2025] [Accepted: 02/12/2025] [Indexed: 03/14/2025] Open
Abstract
BACKGROUND Melanoma is a highly heterogeneous disease, and a deeper molecular classification is essential for improving patient stratification and treatment approaches. Here, we describe the histopathology-driven proteogenomic landscape of 142 treatment-naïve metastatic melanoma samples to uncover molecular subtypes and clinically relevant biomarkers. METHODS We performed an integrative proteogenomic analysis to identify proteomic subtypes, assess the impact of BRAF V600 mutations, and study the molecular profiles and cellular composition of the tumor microenvironment. Clinical and histopathological data were used to support findings related to tissue morphology, disease progression, and patient outcomes. RESULTS Our analysis revealed five distinct proteomic subtypes that integrate immune and stromal microenvironment components and correlate with clinical and histopathological parameters. We demonstrated that BRAF V600-mutated melanomas exhibit biological heterogeneity, where an oncogene-induced senescence-like phenotype is associated with improved survival. This led to a proposed mortality risk-based stratification that may contribute to more personalized treatment strategies. Furthermore, tumor microenvironment composition strongly correlated with disease progression and patient outcomes, highlighting a histopathological connective tissue-to-tumor ratio assessment as a potential decision-making tool. We identified a melanoma-associated SAAV signature linked to extracellular matrix remodeling and SAAV-derived neoantigens as potential targets for anti-tumor immune responses. CONCLUSIONS This study provides a comprehensive stratification of metastatic melanoma, integrating proteogenomic insights with histopathological features. The findings may aid in the development of tailored diagnostic and therapeutic strategies, improving patient management and outcomes.
Collapse
Affiliation(s)
- Magdalena Kuras
- Department of Translational Medicine, Lund University, Skåne University Hospital Malmö, 214 28 Malmö, Sweden; (M.K.); (J.G.); (J.M.); (A.S.); (K.P.)
- Department of Biomedical Engineering, Lund University, 221 00 Lund, Sweden; (P.H.); (I.P.); (J.E.); (Y.S.); (H.L.); (M.R.); (R.A.); (G.M.-V.)
| | - Lazaro Hiram Betancourt
- Department of Translational Medicine, Lund University, Skåne University Hospital Malmö, 214 28 Malmö, Sweden; (M.K.); (J.G.); (J.M.); (A.S.); (K.P.)
- Department of Clinical Sciences Lund, Division of Oncology, Lund University, 221 00 Lund, Sweden; (C.W.); (B.B.); (L.L.); (H.O.)
| | - Runyu Hong
- Institute for Systems Genetics, NYU Grossman School of Medicine, New York, NY 10016, USA; (R.H.); (D.F.)
- Department of Biochemistry and Molecular Pharmacology, NYU Grossman School of Medicine, New York, NY 10016, USA
| | - Leticia Szadai
- Department of Dermatology and Allergology, University of Szeged, 6720 Szeged, Hungary; (L.S.); (I.B.N.)
| | - Jimmy Rodriguez
- Department of Biochemistry and Biophysics, Karolinska Institute, 171 77 Stockholm, Sweden;
| | - Peter Horvatovich
- Department of Biomedical Engineering, Lund University, 221 00 Lund, Sweden; (P.H.); (I.P.); (J.E.); (Y.S.); (H.L.); (M.R.); (R.A.); (G.M.-V.)
- Department of Analytical Biochemistry, Faculty of Science and Engineering, University of Groningen, 9712 CP Groningen, The Netherlands
| | - Indira Pla
- Department of Biomedical Engineering, Lund University, 221 00 Lund, Sweden; (P.H.); (I.P.); (J.E.); (Y.S.); (H.L.); (M.R.); (R.A.); (G.M.-V.)
| | - Jonatan Eriksson
- Department of Biomedical Engineering, Lund University, 221 00 Lund, Sweden; (P.H.); (I.P.); (J.E.); (Y.S.); (H.L.); (M.R.); (R.A.); (G.M.-V.)
| | - Beáta Szeitz
- Division of Oncology, Department of Internal Medicine and Oncology, Semmelweis University, 1085 Budapest, Hungary
| | - Bartłomiej Deszcz
- Department of Biochemistry and Microbiology, Warsaw University of Life Sciences, 02-787 Warsaw, Poland;
| | - Charlotte Welinder
- Department of Clinical Sciences Lund, Division of Oncology, Lund University, 221 00 Lund, Sweden; (C.W.); (B.B.); (L.L.); (H.O.)
| | - Yutaka Sugihara
- Department of Biomedical Engineering, Lund University, 221 00 Lund, Sweden; (P.H.); (I.P.); (J.E.); (Y.S.); (H.L.); (M.R.); (R.A.); (G.M.-V.)
| | - Henrik Ekedahl
- Department of Clinical Sciences Lund, Division of Oncology, Lund University, 221 00 Lund, Sweden; (C.W.); (B.B.); (L.L.); (H.O.)
- SUS University Hospital Lund, 222 42 Lund, Sweden;
| | - Bo Baldetorp
- Department of Clinical Sciences Lund, Division of Oncology, Lund University, 221 00 Lund, Sweden; (C.W.); (B.B.); (L.L.); (H.O.)
| | - Christian Ingvar
- SUS University Hospital Lund, 222 42 Lund, Sweden;
- Department of Surgery, Clinical Sciences, Lund University, SUS, 221 00 Lund, Sweden
| | - Lotta Lundgren
- Department of Clinical Sciences Lund, Division of Oncology, Lund University, 221 00 Lund, Sweden; (C.W.); (B.B.); (L.L.); (H.O.)
- SUS University Hospital Lund, 222 42 Lund, Sweden;
| | - Henrik Lindberg
- Department of Biomedical Engineering, Lund University, 221 00 Lund, Sweden; (P.H.); (I.P.); (J.E.); (Y.S.); (H.L.); (M.R.); (R.A.); (G.M.-V.)
| | - Henriett Oskolas
- Department of Clinical Sciences Lund, Division of Oncology, Lund University, 221 00 Lund, Sweden; (C.W.); (B.B.); (L.L.); (H.O.)
| | - Zsolt Horvath
- Department of Biomedical Engineering, Lund University, 221 00 Lund, Sweden; (P.H.); (I.P.); (J.E.); (Y.S.); (H.L.); (M.R.); (R.A.); (G.M.-V.)
| | - Melinda Rezeli
- Department of Biomedical Engineering, Lund University, 221 00 Lund, Sweden; (P.H.); (I.P.); (J.E.); (Y.S.); (H.L.); (M.R.); (R.A.); (G.M.-V.)
| | - Jeovanis Gil
- Department of Translational Medicine, Lund University, Skåne University Hospital Malmö, 214 28 Malmö, Sweden; (M.K.); (J.G.); (J.M.); (A.S.); (K.P.)
| | - Roger Appelqvist
- Department of Biomedical Engineering, Lund University, 221 00 Lund, Sweden; (P.H.); (I.P.); (J.E.); (Y.S.); (H.L.); (M.R.); (R.A.); (G.M.-V.)
| | - Lajos V. Kemény
- HCEMM-SU Translational Dermatology Research Group, Semmelweis University, 1085 Budapest, Hungary;
- Department of Dermatology, Venereology and Dermatooncology, Faculty of Medicine, Semmelweis University, 1085 Budapest, Hungary
- Department of Physiology, Faculty of Medicine, Semmelweis University, 1085 Budapest, Hungary
- MTA-SE Lendület “Momentum” Dermatology Research Group, Hungarian Academy of Sciences and Semmelweis University, 1085 Budapest, Hungary
| | - Johan Malm
- Department of Translational Medicine, Lund University, Skåne University Hospital Malmö, 214 28 Malmö, Sweden; (M.K.); (J.G.); (J.M.); (A.S.); (K.P.)
| | - Aniel Sanchez
- Department of Translational Medicine, Lund University, Skåne University Hospital Malmö, 214 28 Malmö, Sweden; (M.K.); (J.G.); (J.M.); (A.S.); (K.P.)
| | | | - Krzysztof Pawłowski
- Department of Translational Medicine, Lund University, Skåne University Hospital Malmö, 214 28 Malmö, Sweden; (M.K.); (J.G.); (J.M.); (A.S.); (K.P.)
- Department of Biochemistry and Microbiology, Warsaw University of Life Sciences, 02-787 Warsaw, Poland;
- Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA
| | - Elisabet Wieslander
- Department of Translational Medicine, Lund University, Skåne University Hospital Malmö, 214 28 Malmö, Sweden; (M.K.); (J.G.); (J.M.); (A.S.); (K.P.)
| | - David Fenyö
- Institute for Systems Genetics, NYU Grossman School of Medicine, New York, NY 10016, USA; (R.H.); (D.F.)
- Department of Biochemistry and Molecular Pharmacology, NYU Grossman School of Medicine, New York, NY 10016, USA
| | - Istvan Balazs Nemeth
- Department of Dermatology and Allergology, University of Szeged, 6720 Szeged, Hungary; (L.S.); (I.B.N.)
| | - György Marko-Varga
- Department of Biomedical Engineering, Lund University, 221 00 Lund, Sweden; (P.H.); (I.P.); (J.E.); (Y.S.); (H.L.); (M.R.); (R.A.); (G.M.-V.)
- Chemical Genomics Global Research Lab, Department of Biotechnology, College of Life Science and Biotechnology, Yonsei University, Seoul 03722, Republic of Korea
- 1st Department of Surgery, Tokyo Medical University, Tokyo 160-8402, Japan
| |
Collapse
|
|
4
|
Constâncio V, Lobo J, Sequeira JP, Henrique R, Jerónimo C. Prostate cancer epigenetics - from pathophysiology to clinical application. Nat Rev Urol 2025:10.1038/s41585-024-00991-8. [PMID: 39820138 DOI: 10.1038/s41585-024-00991-8] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Accepted: 12/09/2024] [Indexed: 01/19/2025]
Abstract
Prostate cancer is a multifactorial disease influenced by various molecular features. Over the past decades, epigenetics, which is the study of changes in gene expression without altering the DNA sequence, has been recognized as a major driver of this disease. In the past 50 years, advancements in technological tools to characterize the epigenome have highlighted crucial roles of epigenetic mechanisms throughout the entire spectrum of prostate cancer, from initiation to progression, including localized disease, metastatic dissemination, castration resistance and neuroendocrine transdifferentiation. Substantial advances in the understanding of epigenetic mechanisms in the pathophysiology of prostate cancer have been carried out, but translating preclinical achievements into clinical practice remains challenging. Ongoing research and biomarker-oriented clinical trials are expected to increase the likelihood of successfully integrating epigenetics into prostate cancer clinical management.
Collapse
Affiliation(s)
- Vera Constâncio
- Cancer Biology and Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/CI-IPOP@RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC Raquel Seruca), Porto, Portugal
- Doctoral Program in Biomedical Sciences, ICBAS - School of Medicine & Biomedical Sciences, University of Porto (ICBAS-UP), Porto, Portugal
| | - João Lobo
- Cancer Biology and Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/CI-IPOP@RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC Raquel Seruca), Porto, Portugal
- Department of Pathology, Portuguese Oncology Institute of Porto (IPO Porto)/Porto Comprehensive Cancer Centre Raquel Seruca (Porto.CCC), Porto, Portugal
- Department of Pathology and Molecular Immunology, ICBAS - School of Medicine & Biomedical Sciences, University of Porto, Porto, Portugal
| | - José Pedro Sequeira
- Cancer Biology and Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/CI-IPOP@RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC Raquel Seruca), Porto, Portugal
- Doctoral Program in Biomedical Sciences, ICBAS - School of Medicine & Biomedical Sciences, University of Porto (ICBAS-UP), Porto, Portugal
| | - Rui Henrique
- Cancer Biology and Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/CI-IPOP@RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC Raquel Seruca), Porto, Portugal
- Department of Pathology, Portuguese Oncology Institute of Porto (IPO Porto)/Porto Comprehensive Cancer Centre Raquel Seruca (Porto.CCC), Porto, Portugal
- Department of Pathology and Molecular Immunology, ICBAS - School of Medicine & Biomedical Sciences, University of Porto, Porto, Portugal
| | - Carmen Jerónimo
- Cancer Biology and Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/CI-IPOP@RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC Raquel Seruca), Porto, Portugal.
- Department of Pathology and Molecular Immunology, ICBAS - School of Medicine & Biomedical Sciences, University of Porto, Porto, Portugal.
| |
Collapse
|
|
5
|
Liu X, Li W, Liu Y, Wang X, Shi Q, Yang W, Tu J, Wang Y, Sheng C, Liu N. Discovery of new fungal jumonji H3K27 demethylase inhibitors for the treatment of Cryptococcus neoformans and Candida auris infections. Eur J Med Chem 2025; 281:117028. [PMID: 39536495 DOI: 10.1016/j.ejmech.2024.117028] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 09/01/2024] [Revised: 10/30/2024] [Accepted: 11/02/2024] [Indexed: 11/16/2024]
Abstract
Invasive fungal infections have become a serious public health problem. To tackle the challenges of limited efficacy in antifungal therapy and severe drug resistance, antifungal drugs with new mechanisms of action are urgently needed. Our previous study identified JIB-04 to be an inhibitor of fungal histone demethylase (HDM). To promote target validation and inhibitor design, herein a series of new JIB-04 derivatives were designed and synthesized. After the establishment of structure-activity relationship, compound A4 was identified to possess potent antifungal activity against Cryptococcus neoformans and Candida auris. Compared to lead compound JIB-04, compound A4 was a more potent HDM inhibitor and exhibited better water solubility, virulence factors inhibitory activity and in vivo antifungal potency. Collectively, this study further confirmed that fungal HDMs were potential antifungal targets and compound A4 was a promising antifungal lead compound.
Collapse
Affiliation(s)
- Xin Liu
- The Center for Basic Research and Innovation of Medicine and Pharmacy (MOE), School of Pharmacy, Second Military Medical University (Naval Medical University), 325 Guohe Road, Shanghai, 200433, China
| | - Wang Li
- The Center for Basic Research and Innovation of Medicine and Pharmacy (MOE), School of Pharmacy, Second Military Medical University (Naval Medical University), 325 Guohe Road, Shanghai, 200433, China
| | - Yang Liu
- Department of Pharmacy, NO.971 Hospital of the People's Liberation Army Navy, 22 Minjiang Road, Qingdao, Shandong, 266071, China
| | - Xiaoqing Wang
- The Center for Basic Research and Innovation of Medicine and Pharmacy (MOE), School of Pharmacy, Second Military Medical University (Naval Medical University), 325 Guohe Road, Shanghai, 200433, China
| | - Qiao Shi
- The Center for Basic Research and Innovation of Medicine and Pharmacy (MOE), School of Pharmacy, Second Military Medical University (Naval Medical University), 325 Guohe Road, Shanghai, 200433, China
| | - Wanzhen Yang
- The Center for Basic Research and Innovation of Medicine and Pharmacy (MOE), School of Pharmacy, Second Military Medical University (Naval Medical University), 325 Guohe Road, Shanghai, 200433, China
| | - Jie Tu
- The Center for Basic Research and Innovation of Medicine and Pharmacy (MOE), School of Pharmacy, Second Military Medical University (Naval Medical University), 325 Guohe Road, Shanghai, 200433, China
| | - Yan Wang
- The Center for Basic Research and Innovation of Medicine and Pharmacy (MOE), School of Pharmacy, Second Military Medical University (Naval Medical University), 325 Guohe Road, Shanghai, 200433, China.
| | - Chunquan Sheng
- The Center for Basic Research and Innovation of Medicine and Pharmacy (MOE), School of Pharmacy, Second Military Medical University (Naval Medical University), 325 Guohe Road, Shanghai, 200433, China.
| | - Na Liu
- The Center for Basic Research and Innovation of Medicine and Pharmacy (MOE), School of Pharmacy, Second Military Medical University (Naval Medical University), 325 Guohe Road, Shanghai, 200433, China.
| |
Collapse
|
|
6
|
Nguyen A, Nuñez CG, Tran TA, Girard L, Peyton M, Catalan R, Guerena C, Avila K, Drapkin BJ, Chandra R, Minna JD, Martinez ED. Jumonji histone demethylases are therapeutic targets in small cell lung cancer. Oncogene 2024; 43:2885-2899. [PMID: 39154123 PMCID: PMC11405284 DOI: 10.1038/s41388-024-03125-x] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 01/10/2024] [Revised: 07/30/2024] [Accepted: 08/06/2024] [Indexed: 08/19/2024]
Abstract
Small cell lung cancer (SCLC) is a recalcitrant cancer of neuroendocrine (NE) origin. Changes in therapeutic approaches against SCLC have been lacking over the decades. Here, we use preclinical models to identify a new therapeutic vulnerability in SCLC consisting of the targetable Jumonji lysine demethylase (KDM) family. We show that Jumonji demethylase inhibitors block malignant growth and that etoposide-resistant SCLC cell lines are particularly sensitive to Jumonji inhibition. Mechanistically, small molecule-mediated inhibition of Jumonji KDMs activates endoplasmic reticulum (ER) stress genes, upregulates ER stress signaling, and triggers apoptotic cell death. Furthermore, Jumonji inhibitors decrease protein levels of SCLC NE markers INSM1 and Secretogranin-3 and of driver transcription factors ASCL1 and NEUROD1. Genetic knockdown of KDM4A, a Jumonji demethylase highly expressed in SCLC and a known regulator of ER stress genes, induces ER stress response genes, decreases INSM1, Secretogranin-3, and NEUROD1 and inhibits proliferation of SCLC in vitro and in vivo. Lastly, we demonstrate that two different small molecule Jumonji KDM inhibitors (pan-inhibitor JIB-04 and KDM4 inhibitor SD70) block the growth of SCLC tumor xenografts in vivo. Our study highlights the translational potential of Jumonji KDM inhibitors against SCLC, a clinically feasible approach in light of recently opened clinical trials evaluating this drug class, and establishes KDM4A as a relevant target across SCLC subtypes.
Collapse
Affiliation(s)
- Aiden Nguyen
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
| | - Clarissa G Nuñez
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
| | - Tram Anh Tran
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
| | - Luc Girard
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
- Department of Pharmacology, UT Southwestern Medical Center, Dallas, TX, USA
| | - Michael Peyton
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
| | - Rodrigo Catalan
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
| | - Cristina Guerena
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
| | - Kimberley Avila
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
| | - Benjamin J Drapkin
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
- Department of Internal Medicine, UT Southwestern Medical Center, Dallas, TX, USA
- Simmons Comprehensive Cancer Center, UT Southwestern Medical Center, Dallas, TX, USA
| | - Raghav Chandra
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
- Department of Surgery, UT Southwestern Medical Center, Dallas, TX, USA
| | - John D Minna
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
- Department of Pharmacology, UT Southwestern Medical Center, Dallas, TX, USA
- Department of Internal Medicine, UT Southwestern Medical Center, Dallas, TX, USA
- Simmons Comprehensive Cancer Center, UT Southwestern Medical Center, Dallas, TX, USA
| | - Elisabeth D Martinez
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA.
- Department of Pharmacology, UT Southwestern Medical Center, Dallas, TX, USA.
- Simmons Comprehensive Cancer Center, UT Southwestern Medical Center, Dallas, TX, USA.
| |
Collapse
|
|
7
|
Nakata Y, Nagasawa S, Sera Y, Yamasaki N, Kanai A, Kobatake K, Ueda T, Koizumi M, Manabe I, Kaminuma O, Honda H. PTIP epigenetically regulates DNA damage-induced cell cycle arrest by upregulating PRDM1. Sci Rep 2024; 14:17987. [PMID: 39097652 PMCID: PMC11297997 DOI: 10.1038/s41598-024-68295-w] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 08/31/2023] [Accepted: 07/22/2024] [Indexed: 08/05/2024] Open
Abstract
The genome is constantly exposed to DNA damage from endogenous and exogenous sources. Fine modulation of DNA repair, chromatin remodeling, and transcription factors is necessary for protecting genome integrity, but the precise mechanisms are still largely unclear. We found that after ionizing radiation (IR), global trimethylation of histone H3 at lysine 4 (H3K4me3) was decreased at an early (5 min) post-IR phase but increased at an intermediate (180 min) post-IR phase in both human and mouse hematopoietic cells. We demonstrated that PTIP, a component of the MLL histone methyltransferase complex, is required for H3K4me3 upregulation in the intermediate post-IR phase and promotes cell cycle arrest by epigenetically inducing a cell cycle inhibitor, PRDM1. In addition, we found that PTIP expression is specifically downregulated in acute myeloid leukemia patients. These findings collectively suggest that the PTIP-PRDM1 axis plays an essential role in proper DNA damage response and its deregulation contributes to leukemogenesis.
Collapse
Affiliation(s)
- Yuichiro Nakata
- Department of Systems Medicine, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-Ku, Chiba-Shi, Chiba, 260-8670, Japan.
| | - Shion Nagasawa
- Department of Systems Medicine, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-Ku, Chiba-Shi, Chiba, 260-8670, Japan
| | - Yasuyuki Sera
- Field of Human Disease Models, Major in Advanced Life Sciences and Medicine,, Institute of Laboratory Animals, Tokyo Women's Medical University, 8-1 Kawada-Cho, Shinjuku-Ku, Tokyo, 162-8666, Japan
| | - Norimasa Yamasaki
- Department of Disease Models, Research Institute for Radiation Biology and Medicine, Hiroshima University, 1-2-3 Kasumi, Minami-Ku, Hiroshima, 734-8553, Japan
| | - Akinori Kanai
- Laboratory of Systems Genomics, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, 5-1-5 Kashiwanoha, Kashiwa, Chiba, 277-8561, Japan
| | - Kohei Kobatake
- Department of Urology, Institute of Biomedical and Health Sciences, Hiroshima University, 1-2-3 Kasumi, Minami-Ku, Hiroshima, 734-8553, Japan
| | - Takeshi Ueda
- Department of Biochemistry, Faculty of Medicine, Kindai University, 377-2 Ohnohigashi, Osakasayama-Shi, Osaka, 589-8511, Japan
| | - Miho Koizumi
- Field of Human Disease Models, Major in Advanced Life Sciences and Medicine,, Institute of Laboratory Animals, Tokyo Women's Medical University, 8-1 Kawada-Cho, Shinjuku-Ku, Tokyo, 162-8666, Japan
| | - Ichiro Manabe
- Department of Systems Medicine, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-Ku, Chiba-Shi, Chiba, 260-8670, Japan
| | - Osamu Kaminuma
- Department of Disease Models, Research Institute for Radiation Biology and Medicine, Hiroshima University, 1-2-3 Kasumi, Minami-Ku, Hiroshima, 734-8553, Japan
| | - Hiroaki Honda
- Field of Human Disease Models, Major in Advanced Life Sciences and Medicine,, Institute of Laboratory Animals, Tokyo Women's Medical University, 8-1 Kawada-Cho, Shinjuku-Ku, Tokyo, 162-8666, Japan.
| |
Collapse
|
|
8
|
Dorna D, Kleszcz R, Paluszczak J. Triple Combinations of Histone Lysine Demethylase Inhibitors with PARP1 Inhibitor-Olaparib and Cisplatin Lead to Enhanced Cytotoxic Effects in Head and Neck Cancer Cells. Biomedicines 2024; 12:1359. [PMID: 38927566 PMCID: PMC11201379 DOI: 10.3390/biomedicines12061359] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Key Words] [Grants] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 05/23/2024] [Revised: 06/12/2024] [Accepted: 06/17/2024] [Indexed: 06/28/2024] Open
Abstract
PARP inhibitors are used to treat cancers with a deficient homologous recombination (HR) DNA repair pathway. Interestingly, recent studies revealed that HR repair could be pharmacologically impaired by the inhibition of histone lysine demethylases (KDM). Thus, we investigated whether KDM inhibitors could sensitize head and neck cancer cells, which are usually HR proficient, to PARP inhibition or cisplatin. Therefore, we explored the effects of double combinations of KDM4-6 inhibitors (ML324, CPI-455, GSK-J4, and JIB-04) with olaparib or cisplatin, or their triple combinations with both drugs, on the level of DNA damage and apoptosis. FaDu and SCC-040 cells were treated with individual compounds and their combinations, and cell viability, apoptosis, DNA damage, and gene expression were assessed using the resazurin assay, Annexin V staining, H2A.X activation, and qPCR, respectively. Combinations of KDM inhibitors with cisplatin enhanced cytotoxic effects, unlike combinations with olaparib. Triple combinations of KDM inhibitors with cisplatin and olaparib exhibited the best cytotoxic activity, which was associated with DNA damage accumulation and altered expression of genes associated with apoptosis induction and cell cycle arrest. In conclusion, triple combinations of KDM inhibitors (especially GSK-J4 and JIB-04) with cisplatin and olaparib represent a promising strategy for head and neck cancer treatment.
Collapse
Affiliation(s)
- Dawid Dorna
- Poznan University of Medical Sciences, Doctoral School, Department of Pharmaceutical Biochemistry, 60-806 Poznan, Poland;
| | - Robert Kleszcz
- Poznan University of Medical Sciences, Department of Pharmaceutical Biochemistry, 60-806 Poznan, Poland;
| | - Jarosław Paluszczak
- Poznan University of Medical Sciences, Department of Pharmaceutical Biochemistry, 60-806 Poznan, Poland;
| |
Collapse
|
|
9
|
Tong D, Tang Y, Zhong P. The emerging roles of histone demethylases in cancers. Cancer Metastasis Rev 2024; 43:795-821. [PMID: 38227150 DOI: 10.1007/s10555-023-10160-9] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Journal Information] [Submit a Manuscript] [Subscribe] [Scholar Register] [Received: 04/26/2022] [Accepted: 12/05/2023] [Indexed: 01/17/2024]
Abstract
Modulation of histone methylation status is regarded as an important mechanism of epigenetic regulation and has substantial clinical potential for the therapy of diseases, including cancer and other disorders. The present study aimed to provide a comprehensive introduction to the enzymology of histone demethylases, as well as their cancerous roles, molecular mechanisms, therapeutic possibilities, and challenges for targeting them, in order to advance drug design for clinical therapy and highlight new insight into the mechanisms of these enzymes in cancer. A series of clinical trials have been performed to explore potential roles of histone demethylases in several cancer types. Numerous targeted inhibitors associated with immunotherapy, chemotherapy, radiotherapy, and targeted therapy have been used to exert anticancer functions. Future studies should evaluate the dynamic transformation of histone demethylases leading to carcinogenesis and explore individual therapy.
Collapse
Affiliation(s)
- Dali Tong
- Department of Urological Surgery, Daping Hospital, Army Medical University (Third Military Medical University), Chongqing, 400042, People's Republic of China.
| | - Ying Tang
- Chongqing Key Laboratory of Translational Research for Cancer Metastasis and Individualized Treatment, Chongqing University Cancer Hospital, Chongqing, China.
| | - Peng Zhong
- Department of Pathology, Daping Hospital, Army Medical University (Third Military Medical University), Chongqing, 400042, People's Republic of China.
| |
Collapse
|
|
10
|
Yuan W, Hu J, Wang M, Li G, Lu S, Qiu Y, Liu C, Liu Y. KDM5B promotes metastasis and epithelial-mesenchymal transition via Wnt/β-catenin pathway in squamous cell carcinoma of the head and neck. Mol Carcinog 2024; 63:885-896. [PMID: 38353298 DOI: 10.1002/mc.23695] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 10/03/2023] [Revised: 12/17/2023] [Accepted: 01/22/2024] [Indexed: 04/13/2024]
Abstract
Metastasis determines clinical management decision and restricts the therapeutic efficiency in patients with squamous cell carcinoma of the head and neck (SCCHN). Epigenetic factor KDM5B serves as an oncogene in multiple cancers. However, its role in SCCHN metastasis remains unclear. Our previous study showed that KDM5B is significantly elevated in SCCHN tissue and is positively correlated with metastasis and recurrence. KDM5B overexpression predicted a poor prognosis in both disease-free survival and overall survival, which served as an independent prognostic factor in SCCHN patients. This study further investigates the exact impact of KDM5B in metastasis of SCCHN. We found that KDM5B knockdown significantly inhibits the migration and invasion of SCCHN cells both in vitro and in vivo. On the contrary, forced expression of KDM5B leads to enhanced migration and invasion, accompanied by canonical alterations of epithelial-mesenchymal transition (EMT). Mechanism investigations demonstrated that KDM5B activates Wnt/β-catenin pathway, and inhibition of Wnt/β-catenin pathway via a small molecule inhibitor iCRT-14 partially reverses the enhanced migratory and invasive ability caused by KDM5B in SCCHN cells. Together, our data indicate that KDM5B promotes EMT and metastasis via Wnt/β-catenin pathway in SCCHN, suggesting that KDM5B may be a potential therapeutic target and prognosis biomarker in SCCHN.
Collapse
Affiliation(s)
- Wenhui Yuan
- Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, Hunan, China
- Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Changsha, Hunan, China
- Clinical Research Center for Pharyngolaryngeal Diseases and Voice Disorders in Hunan Province, Changsha, Hunan, China
| | - Junli Hu
- Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, Hunan, China
- Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Changsha, Hunan, China
- Clinical Research Center for Pharyngolaryngeal Diseases and Voice Disorders in Hunan Province, Changsha, Hunan, China
- Department of Otolaryngology Head and Neck Surgery, Yantian District People's Hospital, Shenzhen, Guangdong, China
| | - Mengshu Wang
- Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, Hunan, China
- Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Changsha, Hunan, China
- Clinical Research Center for Pharyngolaryngeal Diseases and Voice Disorders in Hunan Province, Changsha, Hunan, China
| | - Guo Li
- Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, Hunan, China
- Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Changsha, Hunan, China
- Clinical Research Center for Pharyngolaryngeal Diseases and Voice Disorders in Hunan Province, Changsha, Hunan, China
- National Clinical Research Center for Geriatric Disorders (Xiangya Hospital), Changsha, Hunan, China
| | - Shanhong Lu
- Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, Hunan, China
- Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Changsha, Hunan, China
- Clinical Research Center for Pharyngolaryngeal Diseases and Voice Disorders in Hunan Province, Changsha, Hunan, China
- National Clinical Research Center for Geriatric Disorders (Xiangya Hospital), Changsha, Hunan, China
| | - Yuanzheng Qiu
- Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, Hunan, China
- Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Changsha, Hunan, China
- Clinical Research Center for Pharyngolaryngeal Diseases and Voice Disorders in Hunan Province, Changsha, Hunan, China
- National Clinical Research Center for Geriatric Disorders (Xiangya Hospital), Changsha, Hunan, China
| | - Chao Liu
- Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, Hunan, China
- Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Changsha, Hunan, China
- Clinical Research Center for Pharyngolaryngeal Diseases and Voice Disorders in Hunan Province, Changsha, Hunan, China
- National Clinical Research Center for Geriatric Disorders (Xiangya Hospital), Changsha, Hunan, China
| | - Yong Liu
- Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, Hunan, China
- Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Changsha, Hunan, China
- Clinical Research Center for Pharyngolaryngeal Diseases and Voice Disorders in Hunan Province, Changsha, Hunan, China
- National Clinical Research Center for Geriatric Disorders (Xiangya Hospital), Changsha, Hunan, China
| |
Collapse
|
|
11
|
Kim JH, Lee J, Im SS, Kim B, Kim EY, Min HJ, Heo J, Chang EJ, Choi KC, Shin DM, Son J. Glutamine-mediated epigenetic regulation of cFLIP underlies resistance to TRAIL in pancreatic cancer. Exp Mol Med 2024; 56:1013-1026. [PMID: 38684915 PMCID: PMC11058808 DOI: 10.1038/s12276-024-01231-0] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 10/30/2023] [Revised: 03/20/2024] [Accepted: 03/21/2024] [Indexed: 05/02/2024] Open
Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a promising anticancer agent because it kills cancer cells while sparing normal cells. However, many cancers, including pancreatic ductal adenocarcinoma (PDAC), exhibit intrinsic or acquired resistance to TRAIL, and the molecular mechanisms underlying TRAIL resistance in cancers, particularly in PDAC, remain unclear. In this study, we demonstrated that glutamine (Gln) endows PDAC cells with resistance to TRAIL through KDM4C-mediated epigenetic regulation of cFLIP. Inhibition of glutaminolysis significantly reduced the cFLIP level, leading to TRAIL-mediated formation of death-inducing signaling complexes. Overexpression of cFLIP dramatically rescued PDAC cells from TRAIL/Gln deprivation-induced apoptosis. Alpha-Ketoglutarate (aKG) supplementation significantly reversed the decrease in the cFLIP level induced by glutaminolysis inhibition and rescued PDAC cells from TRAIL/Gln deprivation-induced apoptosis. Knockdown of glutamic-oxaloacetic transaminase 2, which facilitates the conversion of oxaloacetate and glutamate into aspartate and aKG, decreased aKG production and the cFLIP level and activated TRAIL-induced apoptosis. AKG-mediated epigenetic regulation was necessary for maintaining a high level of cFLIP. Glutaminolysis inhibition increased the abundance of H3K9me3 in the cFLIP promoter, indicating that Gln-derived aKG production is important for Jumonji-domain histone demethylase (JHDM)-mediated cFLIP regulation. The JHDM KDM4C regulated cFLIP expression by binding to its promoter, and KDM4C knockdown sensitized PDAC cells to TRAIL-induced apoptosis. The present findings suggest that Gln-derived aKG production is required for KDM4C-mediated epigenetic regulation of cFLIP, which leads to resistance to TRAIL.
Collapse
MESH Headings
- Humans
- CASP8 and FADD-Like Apoptosis Regulating Protein/metabolism
- CASP8 and FADD-Like Apoptosis Regulating Protein/genetics
- TNF-Related Apoptosis-Inducing Ligand/metabolism
- Epigenesis, Genetic
- Glutamine/metabolism
- Jumonji Domain-Containing Histone Demethylases/metabolism
- Jumonji Domain-Containing Histone Demethylases/genetics
- Drug Resistance, Neoplasm/genetics
- Pancreatic Neoplasms/metabolism
- Pancreatic Neoplasms/genetics
- Pancreatic Neoplasms/pathology
- Cell Line, Tumor
- Gene Expression Regulation, Neoplastic/drug effects
- Apoptosis/drug effects
- Ketoglutaric Acids/metabolism
- Carcinoma, Pancreatic Ductal/metabolism
- Carcinoma, Pancreatic Ductal/genetics
- Carcinoma, Pancreatic Ductal/pathology
- Aspartate Aminotransferase, Cytoplasmic/metabolism
- Aspartate Aminotransferase, Cytoplasmic/genetics
- Animals
- Promoter Regions, Genetic
Collapse
Affiliation(s)
- Ji Hye Kim
- Department of Biochemistry and Molecular Biology, Brain Korea 21 Project, Asan Medical Center, University of Ulsan College of Medicine, Seoul, 05505, South Korea
| | - Jinyoung Lee
- Department of Biochemistry and Molecular Biology, Brain Korea 21 Project, Asan Medical Center, University of Ulsan College of Medicine, Seoul, 05505, South Korea
| | - Se Seul Im
- Department of Biochemistry and Molecular Biology, Brain Korea 21 Project, Asan Medical Center, University of Ulsan College of Medicine, Seoul, 05505, South Korea
| | - Boyun Kim
- Department of Biochemistry and Molecular Biology, Brain Korea 21 Project, Asan Medical Center, University of Ulsan College of Medicine, Seoul, 05505, South Korea
| | - Eun-Young Kim
- Department of Biochemistry and Molecular Biology, Brain Korea 21 Project, Asan Medical Center, University of Ulsan College of Medicine, Seoul, 05505, South Korea
| | - Hyo-Jin Min
- Department of Biochemistry and Molecular Biology, Brain Korea 21 Project, Asan Medical Center, University of Ulsan College of Medicine, Seoul, 05505, South Korea
| | - Jinbeom Heo
- Department of Cell and Genetic Engineering, Brain Korea 21 Project, Asan Medical Center, University of Ulsan College of Medicine, Seoul, 05505, South Korea
| | - Eun-Ju Chang
- Department of Biochemistry and Molecular Biology, Brain Korea 21 Project, Asan Medical Center, University of Ulsan College of Medicine, Seoul, 05505, South Korea
| | - Kyung-Chul Choi
- Department of Biochemistry and Molecular Biology, Brain Korea 21 Project, Asan Medical Center, University of Ulsan College of Medicine, Seoul, 05505, South Korea
| | - Dong-Myung Shin
- Department of Cell and Genetic Engineering, Brain Korea 21 Project, Asan Medical Center, University of Ulsan College of Medicine, Seoul, 05505, South Korea
| | - Jaekyoung Son
- Department of Biochemistry and Molecular Biology, Brain Korea 21 Project, Asan Medical Center, University of Ulsan College of Medicine, Seoul, 05505, South Korea.
| |
Collapse
|
|
12
|
Macedo-Silva C, Miranda-Gonçalves V, Tavares NT, Barros-Silva D, Lencart J, Lobo J, Oliveira Â, Correia MP, Altucci L, Jerónimo C. Epigenetic regulation of TP53 is involved in prostate cancer radioresistance and DNA damage response signaling. Signal Transduct Target Ther 2023; 8:395. [PMID: 37840069 PMCID: PMC10577134 DOI: 10.1038/s41392-023-01639-6] [Citation(s) in RCA: 2] [Impact Index Per Article: 1.0] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 01/23/2023] [Revised: 08/23/2023] [Accepted: 09/06/2023] [Indexed: 10/17/2023] Open
Abstract
External beam radiotherapy (RT) is a leading first-line therapy for prostate cancer (PCa), and, in recent years, significant advances have been accomplished. However, RT resistance can arise and result in long-term recurrence or disease progression in the worst-case scenario. Thus, making crucial the discovery of new targets for PCa radiosensitization. Herein, we generated a radioresistant PCa cell line, and found p53 to be highly expressed in radioresistant PCa cells, as well as in PCa patients with recurrent/disease progression submitted to RT. Mechanism dissection revealed that RT could promote p53 expression via epigenetic modulation. Specifically, a decrease of H3K27me3 occupancy at TP53 gene promoter, due to increased KDM6B activity, was observed in radioresistant PCa cells. Furthermore, p53 is essential for efficient DNA damage signaling response and cell recovery upon stress induction by prolonged fractionated irradiation. Remarkably, KDM6B inhibition by GSK-J4 significantly decreased p53 expression, consequently attenuating the radioresistant phenotype of PCa cells and hampering in vivo 3D tumor formation. Overall, this work contributes to improve the understanding of p53 as a mediator of signaling transduction in DNA damage repair, as well as the impact of epigenetic targeting for PCa radiosensitization.
Collapse
Grants
- CJ’s Research is funded by Research Center of Portuguese Institute of Porto (BF.CBEG CI-IPOP-27-2016) and EpiParty PI 159-CI-IPOP-152-2021).
- CM-S holds a fellowship grant from UniCampania, Naples, Italy (2019-UNA2CLE-0170010).
- VM-G was funded by P.CCC: Centro Compreensivo de Cancro do Porto” – NORTE-01-0145-FEDER-072678, supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF).
- NTT was funded by P.CCC: Centro Compreensivo de Cancro do Porto” – NORTE-01-0145-FEDER-072678, supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF).
- DB-S holds a fellowship grant from FCT—Fundação para a Ciência e Tecnologia (SFRH/BD/136007/2018).
- MPC was funded by FCT—Fundação para a Ciência e Tecnologia (CEECINST/00091/2018).
- LA’s research is funded by Epi-MS under the VALERE 2019 Program; V:ALERE 2020—“CIRCE”; Campania Regional Government Technology Platform 2038 Lotta alle Patologie Oncologiche iCURE-B21C17000030007; Campania Regional Government FASE2: IDEAL; MIUR, Proof of Concept POC01_00043; POR Campania FSE 2014-2020 ASSE III; PON RI 2014/2020 “Dottorati Innovativi con caratterizzazione ndustrial”; Horizon EU: CAN-SERV BBMRI; EPI-MET MISE 2022; Bando giovani ricercatori D.R. n.834 del 30/09/2022 Università Vanvitelli project: Miranda; National Plan for NRRP Complementary Investments – Law Decree May 6, 2021, n. 59, converted and modified as to Law n. 101/2021Research initiatives for technologies and innovative trajectories in the health and care sectors: project ANTHEM (AdvaNced Technologies for Human-centrEd Medicine).
Collapse
Affiliation(s)
- Catarina Macedo-Silva
- Cancer Biology & Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/ CI-IPOP@ RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO-Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC), R. Dr. António Bernardino de Almeida, 4200-072, Porto, Portugal
- Department of Precision Medicine, University of Campania "Luigi Vanvitelli", 80138, Naples, Italy
| | - Vera Miranda-Gonçalves
- Cancer Biology & Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/ CI-IPOP@ RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO-Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC), R. Dr. António Bernardino de Almeida, 4200-072, Porto, Portugal
- Department of Pathology and Molecular Immunology, ICBAS-School of Medicine & Biomedical Sciences, University of Porto, R. Jorge de Viterbo Ferreira 228, 4050-313, Porto, Portugal
| | - Nuno Tiago Tavares
- Cancer Biology & Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/ CI-IPOP@ RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO-Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC), R. Dr. António Bernardino de Almeida, 4200-072, Porto, Portugal
| | - Daniela Barros-Silva
- Cancer Biology & Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/ CI-IPOP@ RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO-Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC), R. Dr. António Bernardino de Almeida, 4200-072, Porto, Portugal
| | - Joana Lencart
- Medical Physics, Radiobiology and Radiation Protection Group-Research Center of IPO Porto (CI-IPOP)/CI-IPOP@ RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO-Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC), R. Dr. António Bernardino de Almeida, 4200-072, Porto, Portugal
- Department of Medical Physics, Portuguese Oncology Institute of Porto, 4200-072, Porto, Portugal
| | - João Lobo
- Cancer Biology & Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/ CI-IPOP@ RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO-Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC), R. Dr. António Bernardino de Almeida, 4200-072, Porto, Portugal
- Department of Pathology and Molecular Immunology, ICBAS-School of Medicine & Biomedical Sciences, University of Porto, R. Jorge de Viterbo Ferreira 228, 4050-313, Porto, Portugal
- Department of Pathology, Portuguese Oncology Institute of Porto, Porto, Portugal
| | - Ângelo Oliveira
- Department of Radiation Oncology, Portuguese Oncology Institute of Porto, Porto, Portugal
| | - Margareta P Correia
- Cancer Biology & Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/ CI-IPOP@ RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO-Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC), R. Dr. António Bernardino de Almeida, 4200-072, Porto, Portugal
- Department of Pathology and Molecular Immunology, ICBAS-School of Medicine & Biomedical Sciences, University of Porto, R. Jorge de Viterbo Ferreira 228, 4050-313, Porto, Portugal
| | - Lucia Altucci
- Department of Precision Medicine, University of Campania "Luigi Vanvitelli", 80138, Naples, Italy
- BIOGEM, Molecular Biology and Genetics Research Institute, 83100, Avellino, Italy
- IEOS, Institute of Endocrinology and Oncology, 80100, Naples, Italy
| | - Carmen Jerónimo
- Cancer Biology & Epigenetics Group, Research Center of IPO Porto (CI-IPOP)/ CI-IPOP@ RISE (Health Research Network), Portuguese Oncology Institute of Porto (IPO-Porto)/Porto Comprehensive Cancer Center Raquel Seruca (Porto.CCC), R. Dr. António Bernardino de Almeida, 4200-072, Porto, Portugal.
- Department of Pathology and Molecular Immunology, ICBAS-School of Medicine & Biomedical Sciences, University of Porto, R. Jorge de Viterbo Ferreira 228, 4050-313, Porto, Portugal.
| |
Collapse
|
|
13
|
Longbotham JE, Kelly MJS, Fujimori DG. Recognition of Histone H3 Methylation States by the PHD1 Domain of Histone Demethylase KDM5A. ACS Chem Biol 2023; 18:1915-1925. [PMID: 33621062 PMCID: PMC8380758 DOI: 10.1021/acschembio.0c00976] [Citation(s) in RCA: 9] [Impact Index Per Article: 4.5] [Reference Citation Analysis] [Abstract] [MESH Headings] [Grants] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Indexed: 12/25/2022]
Abstract
PHD reader domains are chromatin binding modules often responsible for the recruitment of large protein complexes that contain histone modifying enzymes, chromatin remodelers, and DNA repair machinery. A majority of PHD domains recognize N-terminal residues of histone H3 and are sensitive to the methylation state of Lys4 in histone H3 (H3K4). Histone demethylase KDM5A, an epigenetic eraser enzyme that contains three PHD domains, is often overexpressed in various cancers, and its demethylation activity is allosterically enhanced when its PHD1 domain is bound to the H3 tail. The allosteric regulatory function of PHD1 expands roles of reader domains, suggesting unique features of this chromatin interacting module. Our previous studies determined the H3 binding site of PHD1, although it remains unclear how the H3 tail interacts with the N-terminal residues of PHD1 and how PHD1 discriminates against H3 tails with varying degrees of H3K4 methylation. Here, we have determined the solution structure of apo and H3 bound PHD1. We observe conformational changes occurring in PHD1 in order to accommodate H3, which interestingly binds in a helical conformation. We also observe differential interactions of binding residues with differently methylated H3K4 peptides (me0, me1, me2, or me3), providing a rationale for PHD1's preference for lower methylation states of H3K4. We further assessed the contributions of various H3 interacting residues in the PHD1 domain to the binding of H3 peptides. The structural details of the H3 binding site could provide useful information to aid the development of allosteric small molecule modulators of KDM5A.
Collapse
Affiliation(s)
- James E Longbotham
- Department of Cellular and Molecular Pharmacology, University of California San Francisco, 600 16th Street, Genentech Hall, San Francisco, California 94158, United States
| | - Mark J S Kelly
- Department of Pharmaceutical Chemistry, University of California San Francisco, 600 16th Street, Genentech Hall, San Francisco, California 94158, United States
| | - Danica Galonić Fujimori
- Department of Cellular and Molecular Pharmacology, University of California San Francisco, 600 16th Street, Genentech Hall, San Francisco, California 94158, United States
- Department of Pharmaceutical Chemistry, University of California San Francisco, 600 16th Street, Genentech Hall, San Francisco, California 94158, United States
- Quantitative Biosciences Institute, University of California San Francisco, 1700 Fourth Street, San Francisco, California 94158, United States
| |
Collapse
|
|
14
|
Song YQ, Yang GJ, Ma DL, Wang W, Leung CH. The role and prospect of lysine-specific demethylases in cancer chemoresistance. Med Res Rev 2023; 43:1438-1469. [PMID: 37012609 DOI: 10.1002/med.21955] [Citation(s) in RCA: 5] [Impact Index Per Article: 2.5] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 06/23/2022] [Revised: 02/08/2023] [Accepted: 03/17/2023] [Indexed: 04/05/2023]
Abstract
Histone methylation plays a key function in modulating gene expression, and preserving genome integrity and epigenetic inheritance. However, aberrations of histone methylation are commonly observed in human diseases, especially cancer. Lysine methylation mediated by histone methyltransferases can be reversed by lysine demethylases (KDMs), which remove methyl marks from histone lysine residues. Currently, drug resistance is a main impediment for cancer therapy. KDMs have been found to mediate drug tolerance of many cancers via altering the metabolic profile of cancer cells, upregulating the ratio of cancer stem cells and drug-tolerant genes, and promoting the epithelial-mesenchymal transition and metastatic ability. Moreover, different cancers show distinct oncogenic addictions for KDMs. The abnormal activation or overexpression of KDMs can alter gene expression signatures to enhance cell survival and drug resistance in cancer cells. In this review, we describe the structural features and functions of KDMs, the KDMs preferences of different cancers, and the mechanisms of drug resistance resulting from KDMs. We then survey KDM inhibitors that have been used for combating drug resistance in cancer, and discuss the opportunities and challenges of KDMs as therapeutic targets for cancer drug resistance.
Collapse
Affiliation(s)
- Ying-Qi Song
- State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Macao, China
| | - Guan-Jun Yang
- State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Macao, China
- State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Ningbo University, Ningbo, Zhejiang, China
- Laboratory of Biochemistry and Molecular Biology, School of Marine Sciences, Ningbo University, Ningbo, Zhejiang, China
| | - Dik-Lung Ma
- Department of Chemistry, Hong Kong Baptist University, Hong Kong, China
| | - Wanhe Wang
- Institute of Medical Research, Northwestern Polytechnical University, Xi'an, Shaanxi, China
| | - Chung-Hang Leung
- State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical Sciences, University of Macau, Macao, China
- Department of Biomedical Sciences, Faculty of Health Sciences, University of Macau, Macao, China
| |
Collapse
|
|
15
|
Srivastava R, Singh R, Jauhari S, Lodhi N, Srivastava R. Histone Demethylase Modulation: Epigenetic Strategy to Combat Cancer Progression. EPIGENOMES 2023; 7:epigenomes7020010. [PMID: 37218871 DOI: 10.3390/epigenomes7020010] [Citation(s) in RCA: 2] [Impact Index Per Article: 1.0] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 02/01/2023] [Revised: 05/09/2023] [Accepted: 05/10/2023] [Indexed: 05/24/2023] Open
Abstract
Epigenetic modifications are heritable, reversible changes in histones or the DNA that control gene functions, being exogenous to the genomic sequence itself. Human diseases, particularly cancer, are frequently connected to epigenetic dysregulations. One of them is histone methylation, which is a dynamically reversible and synchronously regulated process that orchestrates the three-dimensional epigenome, nuclear processes of transcription, DNA repair, cell cycle, and epigenetic functions, by adding or removing methylation groups to histones. Over the past few years, reversible histone methylation has become recognized as a crucial regulatory mechanism for the epigenome. With the development of numerous medications that target epigenetic regulators, epigenome-targeted therapy has been used in the treatment of malignancies and has shown meaningful therapeutic potential in preclinical and clinical trials. The present review focuses on the recent advances in our knowledge on the role of histone demethylases in tumor development and modulation, in emphasizing molecular mechanisms that control cancer cell progression. Finally, we emphasize current developments in the advent of new molecular inhibitors that target histone demethylases to regulate cancer progression.
Collapse
Affiliation(s)
- Rashmi Srivastava
- Department of Zoology, Babasaheb Bhimrao Ambedkar University, Lucknow 226025, Uttar Pradesh, India
| | - Rubi Singh
- Department of Hematology, Bioreference Laboratories, Elmwood Park, NJ 07407, USA
| | - Shaurya Jauhari
- Division of Education, Training, and Assessment, Global Education Center, Infosys Limited, Mysuru 570027, Karnataka, India
| | - Niraj Lodhi
- Clinical Research (Research and Development Division) Mirna Analytics LLC, Harlem Bio-Space, New York, NY 10027, USA
| | - Rakesh Srivastava
- Molecular Biology and Microbiology, GenTox Research and Development, Lucknow 226001, Uttar Pradesh, India
| |
Collapse
|
|
16
|
Monte-Serrano E, Morejón-García P, Campillo-Marcos I, Campos-Díaz A, Navarro-Carrasco E, Lazo PA. The pattern of histone H3 epigenetic posttranslational modifications is regulated by the VRK1 chromatin kinase. Epigenetics Chromatin 2023; 16:18. [PMID: 37179361 PMCID: PMC10182654 DOI: 10.1186/s13072-023-00494-7] [Citation(s) in RCA: 7] [Impact Index Per Article: 3.5] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 03/17/2023] [Accepted: 05/06/2023] [Indexed: 05/15/2023] Open
Abstract
BACKGROUND Dynamic chromatin remodeling is associated with changes in the epigenetic pattern of histone acetylations and methylations required for processes based on dynamic chromatin remodeling and implicated in different nuclear functions. These histone epigenetic modifications need to be coordinated, a role that may be mediated by chromatin kinases such as VRK1, which phosphorylates histones H3 and H2A. METHODS The effect of VRK1 depletion and VRK1 inhibitor, VRK-IN-1, on the acetylation and methylation of histone H3 in K4, K9 and K27 was determined under different conditions, arrested or proliferating cells, in A549 lung adenocarcinoma and U2OS osteosarcoma cells. RESULTS Chromatin organization is determined by the phosphorylation pattern of histones mediated by different types of enzymes. We have studied how the VRK1 chromatin kinase can alter the epigenetic posttranslational modifications of histones by using siRNA, a specific inhibitor of this kinase (VRK-IN-1), and of histone acetyl and methyl transferases, as well as histone deacetylase and demethylase. Loss of VRK1 implicated a switch in the state of H3K9 posttranslational modifications. VRK1 depletion/inhibition causes a loss of H3K9 acetylation and facilitates its methylation. This effect is similar to that of the KAT inhibitor C646, and to KDM inhibitors as iadademstat (ORY-1001) or JMJD2 inhibitor. Alternatively, HDAC inhibitors (selisistat, panobinostat, vorinostat) and KMT inhibitors (tazemetostat, chaetocin) have the opposite effect of VRK1 depletion or inhibition, and cause increase of H3K9ac and a decrease of H3K9me3. VRK1 stably interacts with members of these four enzyme families. However, VRK1 can only play a role on these epigenetic modifications by indirect mechanisms in which these epigenetic enzymes are likely targets to be regulated and coordinated by VRK1. CONCLUSIONS The chromatin kinase VRK1 regulates the epigenetic patterns of histone H3 acetylation and methylation in lysines 4, 9 and 27. VRK1 is a master regulator of chromatin organization associated with its specific functions, such as transcription or DNA repair.
Collapse
Affiliation(s)
- Eva Monte-Serrano
- Molecular Mechanisms of Cancer Program, Instituto de Biología Molecular y Celular del Cáncer, Consejo Superior de Investigaciones Científicas (CSIC) - Universidad de Salamanca, 37007, Salamanca, Spain
- Instituto de Investigación Biomédica de Salamanca (IBSAL), Hospital Universitario de Salamanca, 37007, Salamanca, Spain
| | - Patricia Morejón-García
- Molecular Mechanisms of Cancer Program, Instituto de Biología Molecular y Celular del Cáncer, Consejo Superior de Investigaciones Científicas (CSIC) - Universidad de Salamanca, 37007, Salamanca, Spain
- Instituto de Investigación Biomédica de Salamanca (IBSAL), Hospital Universitario de Salamanca, 37007, Salamanca, Spain
| | - Ignacio Campillo-Marcos
- Molecular Mechanisms of Cancer Program, Instituto de Biología Molecular y Celular del Cáncer, Consejo Superior de Investigaciones Científicas (CSIC) - Universidad de Salamanca, 37007, Salamanca, Spain
- Instituto de Investigación Biomédica de Salamanca (IBSAL), Hospital Universitario de Salamanca, 37007, Salamanca, Spain
| | - Aurora Campos-Díaz
- Molecular Mechanisms of Cancer Program, Instituto de Biología Molecular y Celular del Cáncer, Consejo Superior de Investigaciones Científicas (CSIC) - Universidad de Salamanca, 37007, Salamanca, Spain
- Instituto de Investigación Biomédica de Salamanca (IBSAL), Hospital Universitario de Salamanca, 37007, Salamanca, Spain
| | - Elena Navarro-Carrasco
- Molecular Mechanisms of Cancer Program, Instituto de Biología Molecular y Celular del Cáncer, Consejo Superior de Investigaciones Científicas (CSIC) - Universidad de Salamanca, 37007, Salamanca, Spain
- Instituto de Investigación Biomédica de Salamanca (IBSAL), Hospital Universitario de Salamanca, 37007, Salamanca, Spain
| | - Pedro A Lazo
- Molecular Mechanisms of Cancer Program, Instituto de Biología Molecular y Celular del Cáncer, Consejo Superior de Investigaciones Científicas (CSIC) - Universidad de Salamanca, 37007, Salamanca, Spain.
- Instituto de Investigación Biomédica de Salamanca (IBSAL), Hospital Universitario de Salamanca, 37007, Salamanca, Spain.
| |
Collapse
|
|
17
|
Wang XY, Beeraka NM, Xue NN, Yu HM, Yang Y, Liu MX, Nikolenko VN, Liu JQ, Zhao D. Identification of a three-gene prognostic signature for radioresistant esophageal squamous cell carcinoma. World J Clin Oncol 2023; 14:13-26. [PMID: 36699628 PMCID: PMC9850665 DOI: 10.5306/wjco.v14.i1.13] [Citation(s) in RCA: 3] [Impact Index Per Article: 1.5] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Download PDF] [Journal Information] [Submit a Manuscript] [Subscribe] [Scholar Register] [Received: 07/25/2022] [Revised: 10/25/2022] [Accepted: 12/06/2022] [Indexed: 01/10/2023] Open
Abstract
BACKGROUND Esophageal squamous cell carcinoma (ESCC) is causing a high mortality rate due to the lack of efficient early prognosis markers and suitable therapeutic regimens. The prognostic role of genes responsible for the acquisition of radioresistance in ESCC has not been fully elucidated.
AIM To establish a prognostic model by studying gene expression patterns pertinent to radioresistance in ESCC patients.
METHODS Datasets were obtained from the Gene Expression Omnibus and The Cancer Genome Atlas databases. The edgeR, a Bioconductor package, was used to analyze mRNA expression between different groups. We screened genes specifically responsible for radioresistance to estimate overall survival. Pearson correlation analysis was performed to confirm whether the expression of those genes correlated with each other. Genes contributing to radioresistance and overall survival were assessed by the multivariate Cox regression model through the calculation of βi and risk score using the following formula: 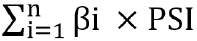 . .
RESULTS We identified three prognostic mRNAs (cathepsin S [CTSS], cluster of differentiation 180 [CD180], and SLP adapter and CSK-interacting membrane protein [SCIMP]) indicative of radioresistance. The expression of the three identified mRNAs was related to each other (r > 0.70 and P < 0.05). As to 1-year and 3-year overall survival prediction, the area under the time-dependent receiver operating characteristic curve of the signature consisting of the three mRNAs was 0.716 and 0.841, respectively. When stratifying patients based on the risk score derived from the signature, the high-risk group exhibited a higher death risk and shorter survival time than the low-risk group (P < 0.0001). Overall survival of the low-risk patients was significantly better than that of the high-risk patients (P = 0.018).
CONCLUSION We have developed a novel three-gene prognostic signature consisting of CTSS, CD180, and SCIMO for ESCC, which may facilitate the prediction of early prognosis of this malignancy.
Collapse
Affiliation(s)
- Xiao-Yan Wang
- Department of Endocrinology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan Province, China
| | - Narasimha M Beeraka
- Department of Radiation Oncology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan Province, China
- Department of Human Anatomy, I. M. Sechenov First Moscow State Medical University, Moscow 119991, Russia
- Department of Pharmaceutical Chemistry, JSS College of Pharmacy, Mysuru 570015, India
| | - Nan-Nan Xue
- Department of Radiation Oncology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan Province, China
| | - Hui-Ming Yu
- Department of Radiation Oncology, Peking University Cancer Hospital & Institute, Beijing 065005, China
| | - Ya Yang
- Department of Radiation Oncology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan Province, China
| | - Mao-Xing Liu
- Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Gastrointestinal Surgery IV, Peking University Cancer Hospital & Institute, Beijing, China
| | - Vladimir N Nikolenko
- Department of Human Anatomy, I. M. Sechenov First Moscow State Medical University, Moscow 119991, Russia
- M.V. Lomonosov Moscow State University, Moscow 119991, Russia
| | - Jun-Qi Liu
- Department of Radiation Oncology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan Province, China
| | - Di Zhao
- Department of Endocrinology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan Province, China
| |
Collapse
|
|
18
|
Jiang Y, Liu L, Yang ZQ. KDM4 Demethylases: Structure, Function, and Inhibitors. ADVANCES IN EXPERIMENTAL MEDICINE AND BIOLOGY 2023; 1433:87-111. [PMID: 37751137 DOI: 10.1007/978-3-031-38176-8_5] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Subscribe] [Scholar Register] [Indexed: 09/27/2023]
Abstract
KDM4 histone demethylases mainly catalyze the removal of methyl marks from H3K9 and H3K36 to epigenetically regulate chromatin structure and gene expression. KDM4 expression is strictly regulated to ensure proper function in a myriad of biological processes, including transcription, cellular proliferation and differentiation, DNA damage repair, immune response, and stem cell self-renewal. Aberrant expression of KDM4 demethylase has been documented in many types of blood and solid tumors, and thus, KDM4s represent promising therapeutic targets. In this chapter, we summarize the current knowledge of the structures and regulatory mechanisms of KDM4 proteins and our understanding of their alterations in human pathological processes with a focus on development and cancer. We also review the reported KDM4 inhibitors and discuss their potential as therapeutic agents.
Collapse
Affiliation(s)
- Yuanyuan Jiang
- Department of Oncology, Karmanos Cancer Institute, Wayne State University, 4100 John R Street, HWCRC 815, Detroit, MI, 48201, USA
| | - Lanxin Liu
- Department of Oncology, Karmanos Cancer Institute, Wayne State University, 4100 John R Street, HWCRC 815, Detroit, MI, 48201, USA
| | - Zeng-Quan Yang
- Department of Oncology, Karmanos Cancer Institute, Wayne State University, 4100 John R Street, HWCRC 815, Detroit, MI, 48201, USA.
| |
Collapse
|
|
19
|
Metzler VM, de Brot S, Haigh DB, Woodcock CL, Lothion-Roy J, Harris AE, Nilsson EM, Ntekim A, Persson JL, Robinson BD, Khani F, Laursen KB, Gudas LJ, Toss MS, Madhusudan S, Rakha E, Heery DM, Rutland CS, Mongan NP, Jeyapalan JN. The KDM5B and KDM1A lysine demethylases cooperate in regulating androgen receptor expression and signalling in prostate cancer. Front Cell Dev Biol 2023; 11:1116424. [PMID: 37152294 PMCID: PMC10154691 DOI: 10.3389/fcell.2023.1116424] [Citation(s) in RCA: 11] [Impact Index Per Article: 5.5] [Reference Citation Analysis] [Abstract] [Key Words] [Grants] [Track Full Text] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 12/05/2022] [Accepted: 04/06/2023] [Indexed: 05/09/2023] Open
Abstract
Histone H3 lysine 4 (H3K4) methylation is key epigenetic mark associated with active transcription and is a substrate for the KDM1A/LSD1 and KDM5B/JARID1B lysine demethylases. Increased expression of KDM1A and KDM5B is implicated in many cancer types, including prostate cancer (PCa). Both KDM1A and KDM5B interact with AR and promote androgen regulated gene expression. For this reason, there is great interested in the development of new therapies targeting KDM1A and KDM5B, particularly in the context of castrate resistant PCa (CRPC), where conventional androgen deprivation therapies and androgen receptor signalling inhibitors are no longer effective. As there is no curative therapy for CRPC, new approaches are urgently required to suppress androgen signalling that prevent, delay or reverse progression to the castrate resistant state. While the contribution of KDM1A to PCa is well established, the exact contribution of KDM5B to PCa is less well understood. However, there is evidence that KDM5B is implicated in numerous pro-oncogenic mechanisms in many different types of cancer, including the hypoxic response, immune evasion and PI3/AKT signalling. Here we elucidate the individual and cooperative functions of KDM1A and KDM5B in PCa. We show that KDM5B mRNA and protein expression is elevated in localised and advanced PCa. We show that the KDM5 inhibitor, CPI-455, impairs androgen regulated transcription and alternative splicing. Consistent with the established role of KDM1A and KDM5B as AR coregulators, we found that individual pharmacologic inhibition of KDM1A and KDM5 by namoline and CPI-455 respectively, impairs androgen regulated transcription. Notably, combined inhibition of KDM1A and KDM5 downregulates AR expression in CRPC cells. Furthermore, combined KDM1A and KDM5 inhibition impairs PCa cell proliferation and invasion more than individual inhibition of KDM1A and KDM5B. Collectively our study has identified individual and cooperative mechanisms involving KDM1A and KDM5 in androgen signalling in PCa. Our findings support the further development of KDM1A and KDM5B inhibitors to treat advanced PCa. Further work is now required to confirm the therapeutic feasibility of combined inhibition of KDM1A and KDM5B as a novel therapeutic strategy for targeting AR positive CRPC.
Collapse
Affiliation(s)
- Veronika M. Metzler
- Biodiscovery Institute, University of Nottingham, Nottingham, United Kingdom
| | - Simone de Brot
- COMPATH, Institute of Animal Pathology, University of Bern, Bern, Switzerland
| | - Daisy B. Haigh
- Biodiscovery Institute, University of Nottingham, Nottingham, United Kingdom
| | - Corinne L. Woodcock
- Biodiscovery Institute, University of Nottingham, Nottingham, United Kingdom
| | | | - Anna E. Harris
- Biodiscovery Institute, University of Nottingham, Nottingham, United Kingdom
| | - Emeli M. Nilsson
- Biodiscovery Institute, University of Nottingham, Nottingham, United Kingdom
| | - Atara Ntekim
- Department of Oncology, University Hospital Ibadan, Ibadan, Nigeria
| | - Jenny L. Persson
- Department of Molecular Biology, Umeå University, Umeå, Sweden
- Department of Biomedical Sciences, Malmö Universitet, Malmö, Sweden
| | - Brian D. Robinson
- Department of Urology, Weill Cornell Medicine, New York, NY, United States
| | - Francesca Khani
- Department of Urology, Weill Cornell Medicine, New York, NY, United States
| | - Kristian B. Laursen
- Department of Pharmacology, Weill Cornell Medicine, New York, NY, United States
| | - Lorraine J. Gudas
- Department of Pharmacology, Weill Cornell Medicine, New York, NY, United States
| | - Michael S. Toss
- Biodiscovery Institute, University of Nottingham, Nottingham, United Kingdom
| | | | - Emad Rakha
- Biodiscovery Institute, University of Nottingham, Nottingham, United Kingdom
| | - David M. Heery
- School of Pharmacy, University of Nottingham, Nottingham, United Kingdom
| | - Catrin S. Rutland
- Biodiscovery Institute, University of Nottingham, Nottingham, United Kingdom
| | - Nigel P. Mongan
- Biodiscovery Institute, University of Nottingham, Nottingham, United Kingdom
- Department of Pharmacology, Weill Cornell Medicine, New York, NY, United States
- *Correspondence: Nigel P. Mongan, , ; Jennie N. Jeyapalan,
| | - Jennie N. Jeyapalan
- Biodiscovery Institute, University of Nottingham, Nottingham, United Kingdom
- *Correspondence: Nigel P. Mongan, , ; Jennie N. Jeyapalan,
| |
Collapse
|
|
20
|
Duan L, Perez RE, Calhoun S, Maki CG. Inhibitors of Jumonji C domain-containing histone lysine demethylases overcome cisplatin and paclitaxel resistance in non-small cell lung cancer through APC/Cdh1-dependent degradation of CtIP and PAF15. Cancer Biol Ther 2022; 23:65-75. [PMID: 35100078 PMCID: PMC8812751 DOI: 10.1080/15384047.2021.2020060] [Citation(s) in RCA: 6] [Impact Index Per Article: 2.0] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Indexed: 12/27/2022] Open
Abstract
The Jumonji C domain-containing family of histone lysine demethylases (Jumonji KDMs) have emerged as promising cancer therapy targets. These enzymes remove methyl groups from various histone lysines and, in turn, regulate processes including chromatin compaction, gene transcription, and DNA repair. Small molecule inhibitors of Jumonji KDMs have shown promise in preclinical studies against non-small cell lung cancer (NSCLC) and other cancers. However, how these inhibitors influence cancer therapy responses and/or DNA repair is incompletely understood. In this study, we established cell line and PDX tumor model systems of cisplatin and paclitaxel-resistant NSCLC. We showed that resistant cells and tumors express high levels of Jumonji-KDMs. Knockdown of individual KDMs or treatment with a pan-Jumonji KDM inhibitor sensitized the cells and tumors to cisplatin and paclitaxel and blocked NSCLC in vivo tumor growth. Mechanistically, we found inhibition of Jumonji-KDMs triggers APC/Cdh1-dependent degradation of CtIP and PAF15, two DNA repair proteins that promote repair of cisplatin and paclitaxel-induced DNA lesions. Knockdown of CtIP and PAF15 sensitized resistant cells to cisplatin, indicating their degradation when Jumonji KDMs are inhibited contributes to cisplatin sensitivity. Our results support the idea that Jumonji-KDMs are a targetable barrier to effective therapy responses in NSCLC. Inhibition of Jumonji KDMs increases therapy (cisplatin/paclitaxel) sensitivity in NSCLC cells, at least in part, by promoting APC/Cdh1-dependent degradation of CtIP and PAF15.
Collapse
Affiliation(s)
- Lei Duan
- Department of Cell & Molecular Medicine, Rush University Medical Center, Chicago, IL, USA
| | - Ricardo E Perez
- Robert H. Lurie Comprehensive Cancer Center of Northwestern University, Chicago, IL, USA
| | - Sarah Calhoun
- Department of Cell & Molecular Medicine, Rush University Medical Center, Chicago, IL, USA
| | - Carl G Maki
- Department of Cell & Molecular Medicine, Rush University Medical Center, Chicago, IL, USA
| |
Collapse
|
|
21
|
Bai J, Shi Z, Wang S, Pan H, Zhang T. MiR-21 and let-7 cooperation in the regulation of lung cancer. Front Oncol 2022; 12:950043. [PMID: 36249072 PMCID: PMC9557158 DOI: 10.3389/fonc.2022.950043] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 05/22/2022] [Accepted: 09/16/2022] [Indexed: 11/30/2022] Open
Abstract
Background Lung cancer occurs and develops as a result of a complicated process involving numerous genes; therefore, single-gene regulation has a limited therapeutic effect. We discovered that miR-21 expression was high in lung cancer tissues and cells, whereas let-7 expression was low, and it is unclear whether their combined regulation would be superior to therapy involving single regulation. The goal of our research was to investigate this situation and the regulatory mechanism that exists between these genes. Methods To regulate the levels of miR-21 and let-7 in these two types of lung cancer cells, we transfected miRNA mimics or inhibitors into A549 and H460 cells. Lung cancer cells were tested for proliferation, apoptosis, migration, and invasion. The results were verified using a Western blot and a qRT-PCR assay. Bioinformatics was used to investigate their potential regulatory pathways, and luciferase assays were used to confirm the binding sites. Results The expression of miR-21 was increased and that of let-7 was decreased in lung cancer tissues and cells compared with paracancerous tissues and normal lung cells (p < 0.01). Tumor cells were inhibited by downregulation of miR-21 and upregulation of let-7, and cooperative regulation showed a better effect. Upregulation of miR-21 and downregulation of let-7 promoted tumor cells, and this tumor-promoting effect was amplified by cooperative regulation. MiR-21 regulated lung cancer cells directly via the Wnt/-catenin pathway, and let-7 exerted its effects via the PLAG1/GDH1 pathway. MiR-21 and let-7 cooperated to regulate lung cancer cells via the K-ras pathway. Conclusions The effect of cooperative regulation of miR-21 and let-7 on lung cancer is greater than that of a single miRNA. MiR-21 and let-7 are important differentially expressed genes in lung cancer that are regulated by the K-ras pathway. As a result, for multigene lung cancer, the cooperative regulation of two miRNAs will provide a new target and direction for lung cancer treatment in the future.
Collapse
|
|
22
|
Wu Q, Young B, Wang Y, Davidoff AM, Rankovic Z, Yang J. Recent Advances with KDM4 Inhibitors and Potential Applications. J Med Chem 2022; 65:9564-9579. [PMID: 35838529 PMCID: PMC9531573 DOI: 10.1021/acs.jmedchem.2c00680] [Citation(s) in RCA: 19] [Impact Index Per Article: 6.3] [Reference Citation Analysis] [Abstract] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Indexed: 11/29/2022]
Abstract
The histone lysine demethylase 4 (KDM4) family plays an important role in regulating gene transcription, DNA repair, and metabolism. The dysregulation of KDM4 functions is associated with many human disorders, including cancer, obesity, and cardiovascular diseases. Selective and potent KDM4 inhibitors may help not only to understand the role of KDM4 in these disorders but also to provide potential therapeutic opportunities. Here, we provide an overview of the field and discuss current status, challenges, and opportunities lying ahead in the development of KDM4-based anticancer therapeutics.
Collapse
Affiliation(s)
- Qiong Wu
- Department of Surgery, St. Jude Children's Research Hospital, Memphis, Tennessee 38105, United States
| | - Brandon Young
- Department of Chemical Biology and Therapeutics, St. Jude Children's Research Hospital, Memphis, Tennessee 38105, United States
| | - Yan Wang
- Department of Geriatrics and Occupational Disease, Qingdao Central Hospital, Qingdao 266044, China
| | - Andrew M Davidoff
- Department of Surgery, St. Jude Children's Research Hospital, Memphis, Tennessee 38105, United States
| | - Zoran Rankovic
- Department of Chemical Biology and Therapeutics, St. Jude Children's Research Hospital, Memphis, Tennessee 38105, United States
| | - Jun Yang
- Department of Surgery, St. Jude Children's Research Hospital, Memphis, Tennessee 38105, United States.,Department of Pathology and Laboratory Medicine, College of Medicine, The University of Tennessee Health Science Center, 930 Madison Avenue, Suite 500, Memphis, Tennessee 38163, United States
| |
Collapse
|
|
23
|
Diverse Functions of KDM5 in Cancer: Transcriptional Repressor or Activator? Cancers (Basel) 2022; 14:cancers14133270. [PMID: 35805040 PMCID: PMC9265395 DOI: 10.3390/cancers14133270] [Citation(s) in RCA: 17] [Impact Index Per Article: 5.7] [Reference Citation Analysis] [Abstract] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 06/04/2022] [Revised: 06/29/2022] [Accepted: 07/02/2022] [Indexed: 11/16/2022] Open
Abstract
Epigenetic modifications are crucial for chromatin remodeling and transcriptional regulation. Post-translational modifications of histones are epigenetic processes that are fine-tuned by writer and eraser enzymes, and the disorganization of these enzymes alters the cellular state, resulting in human diseases. The KDM5 family is an enzymatic family that removes di- and tri-methyl groups (me2 and me3) from lysine 4 of histone H3 (H3K4), and its dysregulation has been implicated in cancer. Although H3K4me3 is an active chromatin marker, KDM5 proteins serve as not only transcriptional repressors but also transcriptional activators in a demethylase-dependent or -independent manner in different contexts. Notably, KDM5 proteins regulate the H3K4 methylation cycle required for active transcription. Here, we review the recent findings regarding the mechanisms of transcriptional regulation mediated by KDM5 in various contexts, with a focus on cancer, and further shed light on the potential of targeting KDM5 for cancer therapy.
Collapse
|
|
24
|
Bayley R, Borel V, Moss RJ, Sweatman E, Ruis P, Ormrod A, Goula A, Mottram RMA, Stanage T, Hewitt G, Saponaro M, Stewart GS, Boulton SJ, Higgs MR. H3K4 methylation by SETD1A/BOD1L facilitates RIF1-dependent NHEJ. Mol Cell 2022; 82:1924-1939.e10. [PMID: 35439434 PMCID: PMC9616806 DOI: 10.1016/j.molcel.2022.03.030] [Citation(s) in RCA: 16] [Impact Index Per Article: 5.3] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 05/10/2021] [Revised: 12/14/2021] [Accepted: 03/23/2022] [Indexed: 12/14/2022]
Abstract
The 53BP1-RIF1-shieldin pathway maintains genome stability by suppressing nucleolytic degradation of DNA ends at double-strand breaks (DSBs). Although RIF1 interacts with damaged chromatin via phospho-53BP1 and facilitates recruitment of the shieldin complex to DSBs, it is unclear whether other regulatory cues contribute to this response. Here, we implicate methylation of histone H3 at lysine 4 by SETD1A-BOD1L in the recruitment of RIF1 to DSBs. Compromising SETD1A or BOD1L expression or deregulating H3K4 methylation allows uncontrolled resection of DNA ends, impairs end-joining of dysfunctional telomeres, and abrogates class switch recombination. Moreover, defects in RIF1 localization to DSBs are evident in patient cells bearing loss-of-function mutations in SETD1A. Loss of SETD1A-dependent RIF1 recruitment in BRCA1-deficient cells restores homologous recombination and leads to resistance to poly(ADP-ribose)polymerase inhibition, reinforcing the clinical relevance of these observations. Mechanistically, RIF1 binds directly to methylated H3K4, facilitating its recruitment to, or stabilization at, DSBs.
Collapse
Affiliation(s)
- Rachel Bayley
- Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham B15 2TT, UK
| | - Valerie Borel
- DSB Repair Metabolism Laboratory, The Francis Crick Institute, Midland Road, London, UK
| | - Rhiannon J Moss
- Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham B15 2TT, UK
| | - Ellie Sweatman
- Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham B15 2TT, UK
| | - Philip Ruis
- DSB Repair Metabolism Laboratory, The Francis Crick Institute, Midland Road, London, UK
| | - Alice Ormrod
- Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham B15 2TT, UK
| | - Amalia Goula
- Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham B15 2TT, UK
| | - Rachel M A Mottram
- Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham B15 2TT, UK
| | - Tyler Stanage
- DSB Repair Metabolism Laboratory, The Francis Crick Institute, Midland Road, London, UK
| | - Graeme Hewitt
- DSB Repair Metabolism Laboratory, The Francis Crick Institute, Midland Road, London, UK
| | - Marco Saponaro
- Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham B15 2TT, UK
| | - Grant S Stewart
- Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham B15 2TT, UK.
| | - Simon J Boulton
- DSB Repair Metabolism Laboratory, The Francis Crick Institute, Midland Road, London, UK.
| | - Martin R Higgs
- Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham B15 2TT, UK.
| |
Collapse
|
|
25
|
Sanchez A, Buck-Koehntop BA, Miller KM. Joining the PARty: PARP Regulation of KDM5A during DNA Repair (and Transcription?). Bioessays 2022; 44:e2200015. [PMID: 35532219 DOI: 10.1002/bies.202200015] [Citation(s) in RCA: 0] [Impact Index Per Article: 0] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 01/19/2022] [Revised: 04/25/2022] [Accepted: 04/28/2022] [Indexed: 11/05/2022]
Abstract
The lysine demethylase KDM5A collaborates with PARP1 and the histone variant macroH2A1.2 to modulate chromatin to promote DNA repair. Indeed, KDM5A engages poly(ADP-ribose) (PAR) chains at damage sites through a previously uncharacterized coiled-coil domain, a novel binding mode for PAR interactions. While KDM5A is a well-known transcriptional regulator, its function in DNA repair is only now emerging. Here we review the molecular mechanisms that regulate this PARP1-macroH2A1.2-KDM5A axis in DNA damage and consider the potential involvement of this pathway in transcription regulation and cancer. Using KDM5A as an example, we discuss how multifunctional chromatin proteins transition between several DNA-based processes, which must be coordinated to protect the integrity of the genome and epigenome. The dysregulation of chromatin and loss of genome integrity that is prevalent in human diseases including cancer may be related and could provide opportunities to target multitasking proteins with these pathways as therapeutic strategies.
Collapse
Affiliation(s)
- Anthony Sanchez
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX, 78712, The University of Texas at Austin, Austin, Texas, USA
| | | | - Kyle M Miller
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX, 78712, The University of Texas at Austin, Austin, Texas, USA.,Livestrong Cancer Institutes, Dell Medical School, The University of Texas at Austin, Austin, Texas, USA
| |
Collapse
|
|
26
|
Son J, Huang S, Zeng Q, Bricker TL, Case JB, Zhou J, Zang R, Liu Z, Chang X, Darling TL, Xu J, Harastani HH, Chen L, Gomez Castro MF, Zhao Y, Kohio HP, Hou G, Fan B, Niu B, Guo R, Rothlauf PW, Bailey AL, Wang X, Shi PY, Martinez ED, Brody SL, Whelan SPJ, Diamond MS, Boon ACM, Li B, Ding S. JIB-04 Has Broad-Spectrum Antiviral Activity and Inhibits SARS-CoV-2 Replication and Coronavirus Pathogenesis. mBio 2022; 13:e0337721. [PMID: 35038906 PMCID: PMC8764536 DOI: 10.1128/mbio.03377-21] [Citation(s) in RCA: 10] [Impact Index Per Article: 3.3] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 11/11/2021] [Accepted: 12/09/2021] [Indexed: 01/18/2023] Open
Abstract
Pathogenic coronaviruses are a major threat to global public health. Here, using a recombinant reporter virus-based compound screening approach, we identified small-molecule inhibitors that potently block the replication of severe acute respiratory syndrome virus 2 (SARS-CoV-2). Among them, JIB-04 inhibited SARS-CoV-2 replication in Vero E6 cells with a 50% effective concentration of 695 nM, with a specificity index of greater than 1,000. JIB-04 showed in vitro antiviral activity in multiple cell types, including primary human bronchial epithelial cells, against several DNA and RNA viruses, including porcine coronavirus transmissible gastroenteritis virus. In an in vivo porcine model of coronavirus infection, administration of JIB-04 reduced virus infection and associated tissue pathology, which resulted in improved weight gain and survival. These results highlight the potential utility of JIB-04 as an antiviral agent against SARS-CoV-2 and other viral pathogens. IMPORTANCE The coronavirus disease 2019 (COVID-19), the disease caused by SARS-CoV-2 infection, is an ongoing public health disaster worldwide. Although several vaccines are available as a preventive measure and the FDA approval of an orally bioavailable drug is on the horizon, there remains a need for developing antivirals against SARS-CoV-2 that could work on the early course of infection. By using infectious reporter viruses, we screened small-molecule inhibitors for antiviral activity against SARS-CoV-2. Among the top hits was JIB-04, a compound previously studied for its anticancer activity. Here, we showed that JIB-04 inhibits the replication of SARS-CoV-2 as well as different DNA and RNA viruses. Furthermore, JIB-04 conferred protection in a porcine model of coronavirus infection, although to a lesser extent when given as therapeutic rather than prophylactic doses. Our findings indicate a limited but still promising utility of JIB-04 as an antiviral agent in the combat against COVID-19 and potentially other viral diseases.
Collapse
Affiliation(s)
- Juhee Son
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA
- Program in Molecular Cell Biology, Washington University in St. Louis, St. Louis, Missouri, USA
| | - Shimeng Huang
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Jiangsu Key Laboratory for Food Quality and Safety-State Key Laboratory Cultivation, Base of Ministry of Science and Technology, Nanjing, China
| | - Qiru Zeng
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Traci L. Bricker
- Department of Medicine, Division of Infectious Diseases, Washington University School of Medicine, St. Louis, Missouri, USA
| | - James Brett Case
- Department of Medicine, Division of Infectious Diseases, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Jinzhu Zhou
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Jiangsu Key Laboratory for Food Quality and Safety-State Key Laboratory Cultivation, Base of Ministry of Science and Technology, Nanjing, China
| | - Ruochen Zang
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA
- Key Laboratory of Marine Drugs, Ministry of Education, Ocean University of China, Qingdao, China
| | - Zhuoming Liu
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Xinjian Chang
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Jiangsu Key Laboratory for Food Quality and Safety-State Key Laboratory Cultivation, Base of Ministry of Science and Technology, Nanjing, China
| | - Tamarand L. Darling
- Department of Medicine, Division of Infectious Diseases, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Jian Xu
- Division of Pulmonary and Critical Care Medicine, Department of Medicine, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Houda H. Harastani
- Department of Medicine, Division of Infectious Diseases, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Lu Chen
- National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, Maryland, USA
| | | | - Yongxiang Zhao
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Jiangsu Key Laboratory for Food Quality and Safety-State Key Laboratory Cultivation, Base of Ministry of Science and Technology, Nanjing, China
| | - Hinissan P. Kohio
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Gaopeng Hou
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Baochao Fan
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Jiangsu Key Laboratory for Food Quality and Safety-State Key Laboratory Cultivation, Base of Ministry of Science and Technology, Nanjing, China
| | - Beibei Niu
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Jiangsu Key Laboratory for Food Quality and Safety-State Key Laboratory Cultivation, Base of Ministry of Science and Technology, Nanjing, China
| | - Rongli Guo
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Jiangsu Key Laboratory for Food Quality and Safety-State Key Laboratory Cultivation, Base of Ministry of Science and Technology, Nanjing, China
| | - Paul W. Rothlauf
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA
- Program in Virology, Harvard Medical School, Boston, Massachusetts, USA
| | - Adam L. Bailey
- Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Xin Wang
- Key Laboratory of Marine Drugs, Ministry of Education, Ocean University of China, Qingdao, China
| | - Pei-Yong Shi
- Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, Texas, USA
| | | | - Steven L. Brody
- Division of Pulmonary and Critical Care Medicine, Department of Medicine, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Sean P. J. Whelan
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Michael S. Diamond
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA
- Department of Medicine, Division of Infectious Diseases, Washington University School of Medicine, St. Louis, Missouri, USA
- Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Adrianus C. M. Boon
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA
- Department of Medicine, Division of Infectious Diseases, Washington University School of Medicine, St. Louis, Missouri, USA
- Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, Missouri, USA
| | - Bin Li
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Jiangsu Key Laboratory for Food Quality and Safety-State Key Laboratory Cultivation, Base of Ministry of Science and Technology, Nanjing, China
| | - Siyuan Ding
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA
| |
Collapse
|
|
27
|
Larionova I, Rakina M, Ivanyuk E, Trushchuk Y, Chernyshova A, Denisov E. Radiotherapy resistance: identifying universal biomarkers for various human cancers. J Cancer Res Clin Oncol 2022; 148:1015-1031. [PMID: 35113235 DOI: 10.1007/s00432-022-03923-4] [Citation(s) in RCA: 29] [Impact Index Per Article: 9.7] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 08/10/2021] [Accepted: 01/12/2022] [Indexed: 12/16/2022]
Abstract
Radiotherapy (RT) is considered as a standard in the treatment of most solid cancers, including glioblastoma, lung, breast, rectal, prostate, colorectal, cervical, esophageal, and head and neck cancers. The main challenge in RT is tumor cell radioresistance associated with a high risk of locoregional relapse and distant metastasis. Despite significant progress in understanding mechanisms of radioresistance, its prediction and overcoming remain unresolved. This review presents the state-of-the-art for the potential universal biomarkers correlated to the radioresistance and poor outcome in different cancers. We describe radioresistance biomarkers functionally attributed to DNA repair, signal transduction, hypoxia, and angiogenesis. We also focus on high throughput genetic and proteomic studies, which revealed a set of molecular biomarkers related to radioresistance. In conclusion, we discuss biomarkers which are overlapped in most several cancers.
Collapse
Affiliation(s)
- Irina Larionova
- Laboratory of Cancer Progression Biology, Cancer Research Institute, Tomsk National Research Medical Center, Russian Academy of Sciences, Tomsk, 634009, Tomsk, Russia.
| | - Militsa Rakina
- Laboratory of Translational Cellular and Molecular Biomedicine, National Research Tomsk State University, Tomsk, 634050, Tomsk, Russia
| | - Elena Ivanyuk
- Laboratory of Cancer Progression Biology, Cancer Research Institute, Tomsk National Research Medical Center, Russian Academy of Sciences, Tomsk, 634009, Tomsk, Russia
| | - Yulia Trushchuk
- Department of Gynecologic Oncology, Cancer Research Institute, Tomsk National Research Medical Center, Russian Academy of Sciences, Tomsk, 634009, Tomsk, Russia
| | - Alena Chernyshova
- Department of Gynecologic Oncology, Cancer Research Institute, Tomsk National Research Medical Center, Russian Academy of Sciences, Tomsk, 634009, Tomsk, Russia
| | - Evgeny Denisov
- Laboratory of Cancer Progression Biology, Cancer Research Institute, Tomsk National Research Medical Center, Russian Academy of Sciences, Tomsk, 634009, Tomsk, Russia
| |
Collapse
|
|
28
|
Zhou B, Zhu Y, Xu W, Zhou Q, Tan L, Zhu L, Chen H, Feng L, Hou T, Wang X, Chen D, Jin H. Hypoxia Stimulates SUMOylation-Dependent Stabilization of KDM5B. Front Cell Dev Biol 2022; 9:741736. [PMID: 34977006 PMCID: PMC8719622 DOI: 10.3389/fcell.2021.741736] [Citation(s) in RCA: 11] [Impact Index Per Article: 3.7] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 07/15/2021] [Accepted: 11/10/2021] [Indexed: 12/25/2022] Open
Abstract
Hypoxia is an important characteristic of the tumor microenvironment. Tumor cells can survive and propagate under the hypoxia stress by activating a series of adaption response. Herein, we found that lysine-specific demethylase 5B (KDM5B) was upregulated in gastric cancer (GC) under hypoxia conditions. The genetic knockdown or chemical inhibition of KDM5B impaired the growth of GC cell adapted to hypoxia. Interestingly, the upregulation of KDM5B in hypoxia response was associated with the SUMOylation of KDM5B. SUMOylation stabilized KDM5B protein by reducing the competitive modification of ubiquitination. Furthermore, the protein inhibitor of activated STAT 4 (PIAS4) was determined as the SUMO E3 ligase, showing increased interaction with KDM5B under hypoxia conditions. The inhibition of KDM5B caused significant downregulation of hypoxia-inducible factor-1α (HIF-1α) protein and target genes under hypoxia. As a result, co-targeting KDM5B significantly improved the antitumor efficacy of antiangiogenic therapy in vivo. Taken together, PIAS4-mediated SUMOylation stabilized KDM5B protein by disturbing ubiquitination-dependent proteasomal degradation to overcome hypoxia stress. Targeting SUMOylation-dependent KDM5B upregulation might be considered when the antiangiogenic therapy was applied in cancer treatment.
Collapse
Affiliation(s)
- Bingluo Zhou
- Laboratory of Cancer Biology, Key Lab of Biotherapy in Zhejiang Province, Cancer Center of Zhejiang University, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
| | - Yiran Zhu
- Laboratory of Cancer Biology, Key Lab of Biotherapy in Zhejiang Province, Cancer Center of Zhejiang University, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
| | - Wenxia Xu
- Laboratory of Cancer Biology, Key Lab of Biotherapy in Zhejiang Province, Cancer Center of Zhejiang University, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
| | - Qiyin Zhou
- Department of Medical Oncology, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
| | - Linghui Tan
- Laboratory of Cancer Biology, Key Lab of Biotherapy in Zhejiang Province, Cancer Center of Zhejiang University, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
| | - Liyuan Zhu
- Laboratory of Cancer Biology, Key Lab of Biotherapy in Zhejiang Province, Cancer Center of Zhejiang University, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
| | - Hui Chen
- Department of Pathology, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
| | - Lifeng Feng
- Laboratory of Cancer Biology, Key Lab of Biotherapy in Zhejiang Province, Cancer Center of Zhejiang University, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
| | - Tianlun Hou
- Department of Clinical Medicine, Wenzhou Medical University, Wenzhou, China
| | - Xian Wang
- Department of Medical Oncology, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
| | - Dingwei Chen
- Department of General Surgery, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
| | - Hongchuan Jin
- Laboratory of Cancer Biology, Key Lab of Biotherapy in Zhejiang Province, Cancer Center of Zhejiang University, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
| |
Collapse
|
|
29
|
Tran TA, Zhang QJ, Wang L, Gonzales C, Girard L, May H, Gillette T, Liu ZP, Martinez ED. Inhibition of Jumonji demethylases reprograms severe dilated cardiomyopathy and prolongs survival. J Biol Chem 2021; 298:101515. [PMID: 34933013 PMCID: PMC8803621 DOI: 10.1016/j.jbc.2021.101515] [Citation(s) in RCA: 4] [Impact Index Per Article: 1.0] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 12/03/2021] [Accepted: 12/08/2021] [Indexed: 12/26/2022] Open
Abstract
Hypertrophic/dilated cardiomyopathy, often a prequel to heart failure, is accompanied by maladaptive transcriptional changes that contribute to arrythmias and contractile misfunction. Transgenic mice constitutively expressing high levels of calcineurin are known to develop extreme heart hypertrophy, which progresses to dilated cardiomyopathy, and to die several weeks after birth. Here, we characterized aberrant transcriptional and epigenetic pathways in this mouse model and established a pharmacological approach to treat established cardiomyopathy. We found that H3K4me3 (trimethyl histone 3 lysine 4) and H3K9me3 (trimethyl histone 3 lysine 9) Jumonji histone demethylases are markedly increased at the protein level and show enhanced enzymatic activity in diseased hearts. These epigenetic regulators continued to increase with time, further affecting cardiac gene expression. Our findings parallel the lower H3K4me3 and H3K9me3 levels seen in human patients. Inhibition of Jumonji demethylase activities in vivo results in lower histone demethylase enzymatic function in the heart and higher histone methylation levels and leads to partial reduction of heart size, reversal of maladaptive transcriptional programs, improved heart function, and prolonged survival. At the molecular level, target genes of transcription factor myocyte enhancer factor 2 are specifically regulated in response to pharmacological or genetic inhibition of Jumonji demethylases. Similar transcriptional reversal of disease-associated genes is seen in a second disease model based on cardiac mechanical overload. Our findings validate pharmacological inhibitors of Jumonji demethylases as potential therapeutics for the treatment of cardiomyopathies across disease models and provide evidence of the reversal of maladaptive transcriptional reprogramming leading to partial restoration of cardiac function. In addition, this study defines pathways of therapeutic resistance upregulated with disease progression.
Collapse
Affiliation(s)
- Tram Anh Tran
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas TX; Department of Pharmacology, UT Southwestern Medical Center, Dallas TX
| | - Qing-Jun Zhang
- Department of Cardiology, UT Southwestern Medical Center, Dallas TX
| | - Lei Wang
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas TX
| | - Christopher Gonzales
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas TX
| | - Luc Girard
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas TX
| | - Herman May
- Department of Cardiology, UT Southwestern Medical Center, Dallas TX
| | - Thomas Gillette
- Department of Cardiology, UT Southwestern Medical Center, Dallas TX
| | - Zhi-Ping Liu
- Department of Cardiology, UT Southwestern Medical Center, Dallas TX.
| | - Elisabeth D Martinez
- Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas TX; Department of Pharmacology, UT Southwestern Medical Center, Dallas TX.
| |
Collapse
|
|
30
|
Carlos-Reyes A, Muñiz-Lino MA, Romero-Garcia S, López-Camarillo C, Hernández-de la Cruz ON. Biological Adaptations of Tumor Cells to Radiation Therapy. Front Oncol 2021; 11:718636. [PMID: 34900673 PMCID: PMC8652287 DOI: 10.3389/fonc.2021.718636] [Citation(s) in RCA: 44] [Impact Index Per Article: 11.0] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 06/01/2021] [Accepted: 10/28/2021] [Indexed: 12/15/2022] Open
Abstract
Radiation therapy has been used worldwide for many decades as a therapeutic regimen for the treatment of different types of cancer. Just over 50% of cancer patients are treated with radiotherapy alone or with other types of antitumor therapy. Radiation can induce different types of cell damage: directly, it can induce DNA single- and double-strand breaks; indirectly, it can induce the formation of free radicals, which can interact with different components of cells, including the genome, promoting structural alterations. During treatment, radiosensitive tumor cells decrease their rate of cell proliferation through cell cycle arrest stimulated by DNA damage. Then, DNA repair mechanisms are turned on to alleviate the damage, but cell death mechanisms are activated if damage persists and cannot be repaired. Interestingly, some cells can evade apoptosis because genome damage triggers the cellular overactivation of some DNA repair pathways. Additionally, some surviving cells exposed to radiation may have alterations in the expression of tumor suppressor genes and oncogenes, enhancing different hallmarks of cancer, such as migration, invasion, and metastasis. The activation of these genetic pathways and other epigenetic and structural cellular changes in the irradiated cells and extracellular factors, such as the tumor microenvironment, is crucial in developing tumor radioresistance. The tumor microenvironment is largely responsible for the poor efficacy of antitumor therapy, tumor relapse, and poor prognosis observed in some patients. In this review, we describe strategies that tumor cells use to respond to radiation stress, adapt, and proliferate after radiotherapy, promoting the appearance of tumor radioresistance. Also, we discuss the clinical impact of radioresistance in patient outcomes. Knowledge of such cellular strategies could help the development of new clinical interventions, increasing the radiosensitization of tumor cells, improving the effectiveness of these therapies, and increasing the survival of patients.
Collapse
Affiliation(s)
- Angeles Carlos-Reyes
- Department of Chronic-Degenerative Diseases, National Institute of Respiratory Diseases “Ismael Cosío Villegas”, Mexico City, Mexico
| | - Marcos A. Muñiz-Lino
- Laboratorio de Patología y Medicina Bucal, Universidad Autónoma Metropolitana Unidad Xochimilco, Mexico City, Mexico
| | - Susana Romero-Garcia
- Department of Chronic-Degenerative Diseases, National Institute of Respiratory Diseases “Ismael Cosío Villegas”, Mexico City, Mexico
| | - César López-Camarillo
- Posgrado en Ciencias Genómicas, Universidad Autónoma de la Ciudad de México, Mexico, Mexico City
| | | |
Collapse
|
|
31
|
Zhao J, Li B, Ren Y, Liang T, Wang J, Zhai S, Zhang X, Zhou P, Zhang X, Pan Y, Gao F, Zhang S, Li L, Yang Y, Deng X, Li X, Chen L, Yang D, Zheng Y. Histone demethylase KDM4A plays an oncogenic role in nasopharyngeal carcinoma by promoting cell migration and invasion. Exp Mol Med 2021; 53:1207-1217. [PMID: 34385569 PMCID: PMC8417295 DOI: 10.1038/s12276-021-00657-0] [Citation(s) in RCA: 10] [Impact Index Per Article: 2.5] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 08/18/2020] [Revised: 06/07/2021] [Accepted: 06/15/2021] [Indexed: 02/07/2023] Open
Abstract
Compelling evidence has indicated the vital role of lysine-specific demethylase 4 A (KDM4A), hypoxia-inducible factor-1α (HIF1α) and the mechanistic target of rapamycin (mTOR) signaling pathway in nasopharyngeal carcinoma (NPC). Therefore, we aimed to investigate whether KDM4A affects NPC progression by regulating the HIF1α/DDIT4/mTOR signaling pathway. First, NPC and adjacent tissue samples were collected, and KDM4A protein expression was examined by immunohistochemistry. Then, the interactions among KDM4A, HIF1α and DDIT4 were assessed. Gain- and loss-of-function approaches were used to alter KDM4A, HIF1α and DDIT4 expression in NPC cells. The mechanism of KDM4A in NPC was evaluated both in vivo and in vitro via RT-qPCR, Western blot analysis, MTT assay, Transwell assay, flow cytometry and tumor formation experiments. KDM4A, HIF1α, and DDIT4 were highly expressed in NPC tissues and cells. Mechanistically, KDM4A inhibited the enrichment of histone H3 lysine 9 trimethylation (H3K9me3) in the HIF1α promoter region and thus inhibited the methylation of HIF1α to promote HIF1α expression, thus upregulating DDIT4 and activating the mTOR signaling pathway. Overexpression of KDM4A, HIF1α, or DDIT4 or activation of the mTOR signaling pathway promoted SUNE1 cell proliferation, migration, and invasion but inhibited apoptosis. KDM4A silencing blocked the mTOR signaling pathway by inhibiting the HIF1α/DDIT4 axis to inhibit the growth of SUNE1 cells in vivo. Collectively, KDM4A silencing could inhibit NPC progression by blocking the activation of the HIF1α/DDIT4/mTOR signaling pathway by increasing H3K9me3, highlighting a promising therapeutic target for NPC.
Collapse
Affiliation(s)
- Jingyi Zhao
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Bingyan Li
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Yongxia Ren
- Radiotherapy Department, Huaihe Hospital of Henan University, Kaifeng, PR China
| | - Tiansong Liang
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Juan Wang
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Suna Zhai
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Xiqian Zhang
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Pengcheng Zhou
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Xiangxian Zhang
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Yuanyuan Pan
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Fangfang Gao
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Sulan Zhang
- Institute of Radiation Therapy and Tumor Critical Care of Zhengzhou University, Zhengzhou, PR China
| | - Liming Li
- Henan Key Laboratory of Molecular Radiotherapy, Zhengzhou, PR China
| | - Yongqiang Yang
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Xiaoyu Deng
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Xiaole Li
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Linhui Chen
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China
| | - Daoke Yang
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China.
| | - Yingjuan Zheng
- Radiotherapy Department, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, PR China.
| |
Collapse
|
|
32
|
Ekstrom TL, Pathoulas NM, Huehls AM, Kanakkanthara A, Karnitz LM. VLX600 Disrupts Homologous Recombination and Synergizes with PARP Inhibitors and Cisplatin by Inhibiting Histone Lysine Demethylases. Mol Cancer Ther 2021; 20:1561-1571. [PMID: 34224364 DOI: 10.1158/1535-7163.mct-20-1099] [Citation(s) in RCA: 7] [Impact Index Per Article: 1.8] [Reference Citation Analysis] [Abstract] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 12/18/2020] [Revised: 02/23/2021] [Accepted: 05/27/2021] [Indexed: 11/16/2022]
Abstract
Tumors with defective homologous recombination (HR) DNA repair are more sensitive to chemotherapies that induce lesions repaired by HR as well as PARP inhibitors (PARPis). However, these therapies have limited activity in HR-proficient cells. Accordingly, agents that disrupt HR may be a means to augment the activities of these therapies in HR-proficient tumors. Here we show that VLX600, a small molecule that has been in a phase I clinical trial, disrupts HR and synergizes with PARPis and platinum compounds in ovarian cancer cells. We further found that VLX600 and other iron chelators disrupt HR, in part, by inhibiting iron-dependent histone lysine demethylases (KDM) family members, thus blocking recruitment of HR repair proteins, including RAD51, to double-strand DNA breaks. Collectively, these findings suggest that pharmacologically targeting KDM family members with VLX600 may be a potential novel strategy to therapeutically induce HR defects in ovarian cancers and correspondingly sensitize them to platinum agents and PARPis, two standard-of-care therapies for ovarian cancer.
Collapse
Affiliation(s)
- Thomas L Ekstrom
- Division of Oncology Research, Mayo Clinic College of Medicine, Mayo Clinic, Rochester, Minnesota
| | - Nicholas M Pathoulas
- Division of Oncology Research, Mayo Clinic College of Medicine, Mayo Clinic, Rochester, Minnesota
| | - Amelia M Huehls
- Division of Oncology Research, Mayo Clinic College of Medicine, Mayo Clinic, Rochester, Minnesota
| | - Arun Kanakkanthara
- Division of Oncology Research, Mayo Clinic College of Medicine, Mayo Clinic, Rochester, Minnesota. .,Department of Molecular Pharmacology and Experimental Therapeutics, Mayo Clinic College of Medicine, Mayo Clinic, Rochester, Minnesota
| | - Larry M Karnitz
- Division of Oncology Research, Mayo Clinic College of Medicine, Mayo Clinic, Rochester, Minnesota. .,Department of Molecular Pharmacology and Experimental Therapeutics, Mayo Clinic College of Medicine, Mayo Clinic, Rochester, Minnesota
| |
Collapse
|
|
33
|
Macedo-Silva C, Benedetti R, Ciardiello F, Cappabianca S, Jerónimo C, Altucci L. Epigenetic mechanisms underlying prostate cancer radioresistance. Clin Epigenetics 2021; 13:125. [PMID: 34103085 PMCID: PMC8186094 DOI: 10.1186/s13148-021-01111-8] [Citation(s) in RCA: 15] [Impact Index Per Article: 3.8] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 03/06/2021] [Accepted: 06/02/2021] [Indexed: 12/24/2022] Open
Abstract
Radiotherapy (RT) is one of the mainstay treatments for prostate cancer (PCa), a highly prevalent neoplasm among males worldwide. About 30% of newly diagnosed PCa patients receive RT with a curative intent. However, biochemical relapse occurs in 20–40% of advanced PCa treated with RT either alone or in combination with adjuvant-hormonal therapy. Epigenetic alterations, frequently associated with molecular variations in PCa, contribute to the acquisition of a radioresistant phenotype. Increased DNA damage repair and cell cycle deregulation decreases radio-response in PCa patients. Moreover, the interplay between epigenome and cell growth pathways is extensively described in published literature. Importantly, as the clinical pattern of PCa ranges from an indolent tumor to an aggressive disease, discovering specific targetable epigenetic molecules able to overcome and predict PCa radioresistance is urgently needed. Currently, histone-deacetylase and DNA-methyltransferase inhibitors are the most studied classes of chromatin-modifying drugs (so-called ‘epidrugs’) within cancer radiosensitization context. Nonetheless, the lack of reliable validation trials is a foremost drawback. This review summarizes the major epigenetically induced changes in radioresistant-like PCa cells and describes recently reported targeted epigenetic therapies in pre-clinical and clinical settings. ![]()
Collapse
Affiliation(s)
- Catarina Macedo-Silva
- Department of Precision Medicine, University of Campania "Luigi Vanvitelli", Vico L. De Crecchio 7, 80138, Naplei, Italy.,Cancer Biology and Epigenetics Group, Research Center at Portuguese Oncology Institute of Porto, F Bdg, 1st Floor, Rua Dr. António Bernardino de Almeida, 4200-072, Porto, Portugal
| | - Rosaria Benedetti
- Department of Precision Medicine, University of Campania "Luigi Vanvitelli", Vico L. De Crecchio 7, 80138, Naplei, Italy
| | - Fortunato Ciardiello
- Department of Precision Medicine, University of Campania "Luigi Vanvitelli", Vico L. De Crecchio 7, 80138, Naplei, Italy
| | - Salvatore Cappabianca
- Department of Precision Medicine, University of Campania "Luigi Vanvitelli", Vico L. De Crecchio 7, 80138, Naplei, Italy
| | - Carmen Jerónimo
- Cancer Biology and Epigenetics Group, Research Center at Portuguese Oncology Institute of Porto, F Bdg, 1st Floor, Rua Dr. António Bernardino de Almeida, 4200-072, Porto, Portugal. .,Department of Pathology and Molecular Immunology at School of Medicine and Biomedical Sciences, University of Porto (ICBAS-UP), Porto, Portugal.
| | - Lucia Altucci
- Department of Precision Medicine, University of Campania "Luigi Vanvitelli", Vico L. De Crecchio 7, 80138, Naplei, Italy.
| |
Collapse
|
|
34
|
Son J, Huang S, Zeng Q, Bricker TL, Case JB, Zhou J, Zang R, Liu Z, Chang X, Harastani HH, Chen L, Castro MFG, Zhao Y, Kohio HP, Hou G, Fan B, Niu B, Guo R, Rothlauf PW, Bailey AL, Wang X, Shi PY, Martinez ED, Whelan SP, Diamond MS, Boon AC, Li B, Ding S. JIB-04 has broad-spectrum antiviral activity and inhibits SARS-CoV-2 replication and coronavirus pathogenesis. BIORXIV : THE PREPRINT SERVER FOR BIOLOGY 2021:2020.09.24.312165. [PMID: 32995798 PMCID: PMC7523209 DOI: 10.1101/2020.09.24.312165] [Citation(s) in RCA: 7] [Impact Index Per Article: 1.8] [Reference Citation Analysis] [Abstract] [Grants] [Track Full Text] [Download PDF] [Figures] [Subscribe] [Scholar Register] [Indexed: 02/07/2023]
Abstract
Pathogenic coronaviruses represent a major threat to global public health. Here, using a recombinant reporter virus-based compound screening approach, we identified several small-molecule inhibitors that potently block the replication of the newly emerged severe acute respiratory syndrome virus 2 (SARS-CoV-2). Among them, JIB-04 inhibited SARS-CoV-2 replication in Vero E6 cells with an EC50 of 695 nM, with a specificity index of greater than 1,000. JIB-04 showed in vitro antiviral activity in multiple cell types against several DNA and RNA viruses, including porcine coronavirus transmissible gastroenteritis virus. In an in vivo porcine model of coronavirus infection, administration of JIB-04 reduced virus infection and associated tissue pathology, which resulted in improved weight gain and survival. These results highlight the potential utility of JIB-04 as an antiviral agent against SARS-CoV-2 and other viral pathogens.
Collapse
Affiliation(s)
- Juhee Son
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO, USA
- Program in Molecular Cell Biology, Washington University School of Medicine, St. Louis, MO, USA
| | - Shimeng Huang
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Nanjing, China
| | - Qiru Zeng
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO, USA
| | - Traci L. Bricker
- Department of Medicine, Division of Infectious Diseases, Washington University School of Medicine, St. Louis, MO, USA
| | - James Brett Case
- Department of Medicine, Division of Infectious Diseases, Washington University School of Medicine, St. Louis, MO, USA
| | - Jinzhu Zhou
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Nanjing, China
| | - Ruochen Zang
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO, USA
- Key Laboratory of Marine Drugs, Ministry of Education, Ocean University of China, Qingdao, China
| | - Zhuoming Liu
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO, USA
| | - Xinjian Chang
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Nanjing, China
| | - Houda H. Harastani
- Department of Medicine, Division of Infectious Diseases, Washington University School of Medicine, St. Louis, MO, USA
| | - Lu Chen
- National Center for Advancing Translational Sciences, National Institutes of Health, Rockville, MD, USA
| | | | - Yongxiang Zhao
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Nanjing, China
| | - Hinissan P. Kohio
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO, USA
| | - Gaopeng Hou
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO, USA
| | - Baochao Fan
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Nanjing, China
| | - Beibei Niu
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Nanjing, China
| | - Rongli Guo
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Nanjing, China
| | - Paul W. Rothlauf
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO, USA
- Program in Virology, Harvard Medical School, Boston, MA, USA
| | - Adam L. Bailey
- Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO, USA
| | - Xin Wang
- Key Laboratory of Marine Drugs, Ministry of Education, Ocean University of China, Qingdao, China
| | - Pei-Yong Shi
- Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston TX, USA
| | | | - Sean P.J. Whelan
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO, USA
| | - Michael S. Diamond
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO, USA
- Department of Medicine, Division of Infectious Diseases, Washington University School of Medicine, St. Louis, MO, USA
- Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO, USA
| | - Adrianus C.M. Boon
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO, USA
- Department of Medicine, Division of Infectious Diseases, Washington University School of Medicine, St. Louis, MO, USA
- Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, MO, USA
| | - Bin Li
- Institute of Veterinary Medicine, Jiangsu Academy of Agricultural Sciences, Nanjing, China
| | - Siyuan Ding
- Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO, USA
| |
Collapse
|
|
35
|
Gaillard S, Charasson V, Ribeyre C, Salifou K, Pillaire MJ, Hoffmann JS, Constantinou A, Trouche D, Vandromme M. KDM5A and KDM5B histone-demethylases contribute to HU-induced replication stress response and tolerance. Biol Open 2021; 10:268370. [PMID: 34184733 PMCID: PMC8181900 DOI: 10.1242/bio.057729] [Citation(s) in RCA: 4] [Impact Index Per Article: 1.0] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 11/06/2020] [Accepted: 04/20/2021] [Indexed: 12/25/2022] Open
Abstract
KDM5A and KDM5B histone-demethylases are overexpressed in many cancers and have been involved in drug tolerance. Here, we describe that KDM5A, together with KDM5B, contribute to replication stress (RS) response and tolerance. First, they positively regulate RRM2, the regulatory subunit of ribonucleotide reductase. Second, they are required for optimal levels of activated Chk1, a major player of the intra-S phase checkpoint that protects cells from RS. We also found that KDM5A is enriched at ongoing replication forks and associates with both PCNA and Chk1. Because RRM2 is a major determinant of replication stress tolerance, we developed cells resistant to HU, and show that KDM5A/B proteins are required for both RRM2 overexpression and tolerance to HU. Altogether, our results indicate that KDM5A/B are major players of RS management. They also show that drugs targeting the enzymatic activity of KDM5 proteins may not affect all cancer-related consequences of KDM5A/B overexpression.
Collapse
Affiliation(s)
- Solenne Gaillard
- MCD, Centre de Biologie Integrative (CBI), University of Toulouse, CNRS, UPS, 31062 Toulouse, France
| | - Virginie Charasson
- MCD, Centre de Biologie Integrative (CBI), University of Toulouse, CNRS, UPS, 31062 Toulouse, France
| | - Cyril Ribeyre
- Institut de Génétique Humaine, CNRS, Université de Montpellier, Montpellier, France
| | - Kader Salifou
- Institut de Génétique Humaine, CNRS, Université de Montpellier, Montpellier, France
| | - Marie-Jeanne Pillaire
- Cancer Research Center of Toulouse, INSERM U1037, CNRS ERL5294, University of Toulouse 3, 31037 Toulouse, France
| | - Jean-Sebastien Hoffmann
- Laboratoire de Pathologie, Institut Universitaire du Cancer-Toulouse, Oncopole, 1 avenue Irène-Joliot-Curie, 31059 Toulouse cedex, France
| | - Angelos Constantinou
- Institut de Génétique Humaine, CNRS, Université de Montpellier, Montpellier, France
| | - Didier Trouche
- MCD, Centre de Biologie Integrative (CBI), University of Toulouse, CNRS, UPS, 31062 Toulouse, France
| | - Marie Vandromme
- MCD, Centre de Biologie Integrative (CBI), University of Toulouse, CNRS, UPS, 31062 Toulouse, France
| |
Collapse
|
|
36
|
Kumbhar R, Sanchez A, Perren J, Gong F, Corujo D, Medina F, Devanathan SK, Xhemalce B, Matouschek A, Buschbeck M, Buck-Koehntop BA, Miller KM. Poly(ADP-ribose) binding and macroH2A mediate recruitment and functions of KDM5A at DNA lesions. J Cell Biol 2021; 220:212163. [PMID: 34003252 PMCID: PMC8135068 DOI: 10.1083/jcb.202006149] [Citation(s) in RCA: 11] [Impact Index Per Article: 2.8] [Reference Citation Analysis] [Abstract] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 06/24/2020] [Revised: 03/15/2021] [Accepted: 04/12/2021] [Indexed: 12/13/2022] Open
Abstract
The histone demethylase KDM5A erases histone H3 lysine 4 methylation, which is involved in transcription and DNA damage responses (DDRs). While DDR functions of KDM5A have been identified, how KDM5A recognizes DNA lesion sites within chromatin is unknown. Here, we identify two factors that act upstream of KDM5A to promote its association with DNA damage sites. We have identified a noncanonical poly(ADP-ribose) (PAR)–binding region unique to KDM5A. Loss of the PAR-binding region or treatment with PAR polymerase (PARP) inhibitors (PARPi’s) blocks KDM5A–PAR interactions and DNA repair functions of KDM5A. The histone variant macroH2A1.2 is also specifically required for KDM5A recruitment and function at DNA damage sites, including homology-directed repair of DNA double-strand breaks and repression of transcription at DNA breaks. Overall, this work reveals the importance of PAR binding and macroH2A1.2 in KDM5A recognition of DNA lesion sites that drive transcriptional and repair activities at DNA breaks within chromatin that are essential for maintaining genome integrity.
Collapse
Affiliation(s)
- Ramhari Kumbhar
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX.,Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX
| | - Anthony Sanchez
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX.,Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX
| | - Jullian Perren
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX.,Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX
| | - Fade Gong
- Department of Biochemistry & Molecular Biology, Baylor College of Medicine, Houston, TX
| | - David Corujo
- Cancer and Leukemia Epigenetics and Biology Program, Josep Carreras Leukaemia Cancer Institute, Barcelona, Spain
| | - Frank Medina
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX.,Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX
| | - Sravan K Devanathan
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX.,Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX
| | - Blerta Xhemalce
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX.,Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX.,Livestrong Cancer Institutes, Dell Medical School, The University of Texas at Austin, Austin, TX
| | - Andreas Matouschek
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX.,Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX
| | - Marcus Buschbeck
- Cancer and Leukemia Epigenetics and Biology Program, Josep Carreras Leukaemia Cancer Institute, Barcelona, Spain.,Program for Predictive and Personalized Medicine of Cancer, Germans Trias i Pujol Research Institute, Badalona, Spain
| | | | - Kyle M Miller
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX.,Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX.,Livestrong Cancer Institutes, Dell Medical School, The University of Texas at Austin, Austin, TX
| |
Collapse
|
|
37
|
Bhan A, Ansari KI, Chen MY, Jandial R. Inhibition of Jumonji Histone Demethylases Selectively Suppresses HER2 + Breast Leptomeningeal Carcinomatosis Growth via Inhibition of GMCSF Expression. Cancer Res 2021; 81:3200-3214. [PMID: 33941612 DOI: 10.1158/0008-5472.can-20-3317] [Citation(s) in RCA: 12] [Impact Index Per Article: 3.0] [Reference Citation Analysis] [Abstract] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 09/30/2020] [Revised: 03/17/2021] [Accepted: 04/28/2021] [Indexed: 11/16/2022]
Abstract
HER2+ breast leptomeningeal carcinomatosis (HER2+ LC) occurs when tumor cells spread to cerebrospinal fluid-containing leptomeninges surrounding the brain and spinal cord, a complication with a dire prognosis. HER2+ LC remains incurable, with few treatment options. Currently, much effort is devoted toward development of therapies that target mutations. However, targeting epigenetic or transcriptional states of HER2+ LC tumors might efficiently target HER2+ LC growth via inhibition of oncogenic signaling; this approach remains promising but is less explored. To test this possibility, we established primary HER2+ LC (Lepto) cell lines from nodular HER2+ LC tissues. These lines are phenotypically CD326+CD49f-, confirming that they are derived from HER2+ LC tumors, and express surface CD44+CD24-, a cancer stem cell (CSC) phenotype. Like CSCs, Lepto lines showed greater drug resistance and more aggressive behavior compared with other HER2+ breast cancer lines in vitro and in vivo. Interestingly, the three Lepto lines overexpressed Jumonji domain-containing histone lysine demethylases KDM4A/4C. Treatment with JIB04, a selective inhibitor of Jumonji demethylases, or genetic loss of function of KDM4A/4C induced apoptosis and cell-cycle arrest and reduced Lepto cell viability, tumorsphere formation, regrowth, and invasion in vitro. JIB04 treatment of patient-derived xenograft mouse models in vivo reduced HER2+ LC tumor growth and prolonged animal survival. Mechanistically, KDM4A/4C inhibition downregulated GMCSF expression and prevented GMCSF-dependent Lepto cell proliferation. Collectively, these results establish KDM4A/4C as a viable therapeutic target in HER2+ LC and spotlight the benefits of targeting the tumorigenic transcriptional network. SIGNIFICANCE: HER2+ LC tumors overexpress KDM4A/4C and are sensitive to the Jumonji demethylase inhibitor JIB04, which reduces the viability of primary HER2+ LC cells and increases survival in mouse models.
Collapse
Affiliation(s)
- Arunoday Bhan
- Division of Neurosurgery, Beckman Research Institute, City of Hope, Duarte, California
| | - Khairul I Ansari
- Division of Neurosurgery, Beckman Research Institute, City of Hope, Duarte, California.,Celcuity, Minneapolis, Minnesota
| | - Mike Y Chen
- Division of Neurosurgery, Beckman Research Institute, City of Hope, Duarte, California
| | - Rahul Jandial
- Division of Neurosurgery, Beckman Research Institute, City of Hope, Duarte, California.
| |
Collapse
|
|
38
|
Di Nisio E, Lupo G, Licursi V, Negri R. The Role of Histone Lysine Methylation in the Response of Mammalian Cells to Ionizing Radiation. Front Genet 2021; 12:639602. [PMID: 33859667 PMCID: PMC8042281 DOI: 10.3389/fgene.2021.639602] [Citation(s) in RCA: 24] [Impact Index Per Article: 6.0] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 12/09/2020] [Accepted: 03/11/2021] [Indexed: 12/20/2022] Open
Abstract
Eukaryotic genomes are wrapped around nucleosomes and organized into different levels of chromatin structure. Chromatin organization has a crucial role in regulating all cellular processes involving DNA-protein interactions, such as DNA transcription, replication, recombination and repair. Histone post-translational modifications (HPTMs) have a prominent role in chromatin regulation, acting as a sophisticated molecular code, which is interpreted by HPTM-specific effectors. Here, we review the role of histone lysine methylation changes in regulating the response to radiation-induced genotoxic damage in mammalian cells. We also discuss the role of histone methyltransferases (HMTs) and histone demethylases (HDMs) and the effects of the modulation of their expression and/or the pharmacological inhibition of their activity on the radio-sensitivity of different cell lines. Finally, we provide a bioinformatic analysis of published datasets showing how the mRNA levels of known HMTs and HDMs are modulated in different cell lines by exposure to different irradiation conditions.
Collapse
Affiliation(s)
- Elena Di Nisio
- Department of Biology and Biotechnology Charles Darwin, Sapienza University of Rome, Rome, Italy
| | - Giuseppe Lupo
- Department of Biology and Biotechnology Charles Darwin, Sapienza University of Rome, Rome, Italy
| | - Valerio Licursi
- Department of Biology and Biotechnology Charles Darwin, Sapienza University of Rome, Rome, Italy
| | - Rodolfo Negri
- Department of Biology and Biotechnology Charles Darwin, Sapienza University of Rome, Rome, Italy.,Institute of Molecular Biology and Pathology, National Research Counsil (IBPM-CNR), Rome, Italy
| |
Collapse
|
|
39
|
Osrodek M, Wozniak M. Targeting Genome Stability in Melanoma-A New Approach to an Old Field. Int J Mol Sci 2021; 22:3485. [PMID: 33800547 PMCID: PMC8036881 DOI: 10.3390/ijms22073485] [Citation(s) in RCA: 2] [Impact Index Per Article: 0.5] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 03/04/2021] [Revised: 03/24/2021] [Accepted: 03/25/2021] [Indexed: 02/07/2023] Open
Abstract
Despite recent groundbreaking advances in the treatment of cutaneous melanoma, it remains one of the most treatment-resistant malignancies. Due to resistance to conventional chemotherapy, the therapeutic focus has shifted away from aiming at melanoma genome stability in favor of molecularly targeted therapies. Inhibitors of the RAS/RAF/MEK/ERK (MAPK) pathway significantly slow disease progression. However, long-term clinical benefit is rare due to rapid development of drug resistance. In contrast, immune checkpoint inhibitors provide exceptionally durable responses, but only in a limited number of patients. It has been increasingly recognized that melanoma cells rely on efficient DNA repair for survival upon drug treatment, and that genome instability increases the efficacy of both MAPK inhibitors and immunotherapy. In this review, we discuss recent developments in the field of melanoma research which indicate that targeting genome stability of melanoma cells may serve as a powerful strategy to maximize the efficacy of currently available therapeutics.
Collapse
Affiliation(s)
| | - Michal Wozniak
- Department of Molecular Biology of Cancer, Medical University of Lodz, 92-215 Lodz, Poland;
| |
Collapse
|
|
40
|
Cabrera-Licona A, Pérez-Añorve IX, Flores-Fortis M, Moral-Hernández OD, González-de la Rosa CH, Suárez-Sánchez R, Chávez-Saldaña M, Aréchaga-Ocampo E. Deciphering the epigenetic network in cancer radioresistance. Radiother Oncol 2021; 159:48-59. [PMID: 33741468 DOI: 10.1016/j.radonc.2021.03.012] [Citation(s) in RCA: 11] [Impact Index Per Article: 2.8] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 08/28/2020] [Revised: 02/15/2021] [Accepted: 03/09/2021] [Indexed: 12/16/2022]
Abstract
Radiotherapy, in addition to surgery and systemic chemotherapy, remains the core of the current clinical management of cancer. Radioresistance is one of the major causes of disease progression and mortality in cancer; therefore, it is a significant challenge in the treatment of locally advanced, recurrent and metastatic cancer. Epigenetic mechanisms that control hallmarks of cancer have a key role in the development of radiation resistance of cancer cells. Recent advances in DNA methylation, histone modification, chromatin remodeling and non-coding RNAs identified in the control of signal transduction pathways in cancer and cancer stem cells have provided even greater promise in the improvement of understanding cancer radioresistance. Many epigenetic drugs that target epigenetic enzymes revert the radioresistant phenotypes decreasing the possibility that resistant cancer cells will develop refractory tumors to radiotherapy. Epigenetic profiles identified as regulators of DNA damage repair, hypoxia, cell survival, apoptosis and invasion are determinants in the development of tumor radioresistance; hence, they also are promising in personalized medicine to develop novel targeted therapies or biomarkers to follow-up the effectiveness of radiotherapy. Now, it is clear that radiotherapy can influence a complex epigenetic network for transcriptional reprogramming, enabling the cells to adapt and avoid the effect of radiotherapy. This review aims to highlight the epigenetic modifications identified in cancer radioresistance and to discuss approaches to disable epigenetic networks to increase the sensitivity and specificity of radiotherapy.
Collapse
Affiliation(s)
- Ariana Cabrera-Licona
- Departamento de Ciencias Naturales, Unidad Cuajimalpa, Universidad Autonoma Metropolitana, Ciudad de Mexico, Mexico; Posgrado en Ciencias Naturales e Ingenieria, Unidad Cuajimalpa, Universidad Autonoma Metropolitana, Ciudad de Mexico, Mexico
| | - Isidro X Pérez-Añorve
- Departamento de Ciencias Naturales, Unidad Cuajimalpa, Universidad Autonoma Metropolitana, Ciudad de Mexico, Mexico
| | - Mauricio Flores-Fortis
- Departamento de Ciencias Naturales, Unidad Cuajimalpa, Universidad Autonoma Metropolitana, Ciudad de Mexico, Mexico; Posgrado en Ciencias Naturales e Ingenieria, Unidad Cuajimalpa, Universidad Autonoma Metropolitana, Ciudad de Mexico, Mexico
| | - Oscar Del Moral-Hernández
- Laboratorio de Virologia y Epigenetica del Cancer, Facultad de Ciencias Quimico Biologicas, Universidad Autonoma de Guerrero, Chilpancingo, Mexico
| | | | - Rocio Suárez-Sánchez
- Laboratorio de Medicina Genomica, Departamento de Genetica, Instituto Nacional de Rehabilitacion LGII, Ciudad de Mexico, Mexico
| | - Margarita Chávez-Saldaña
- Laboratorio de Biologia de la Reproduccion, Instituto Nacional de Pediatria, Ciudad de Mexico, Mexico
| | - Elena Aréchaga-Ocampo
- Departamento de Ciencias Naturales, Unidad Cuajimalpa, Universidad Autonoma Metropolitana, Ciudad de Mexico, Mexico.
| |
Collapse
|
|
41
|
Khaliq M, Manikkam M, Martinez ED, Fallahi-Sichani M. Epigenetic modulation reveals differentiation state specificity of oncogene addiction. Nat Commun 2021; 12:1536. [PMID: 33750776 PMCID: PMC7943789 DOI: 10.1038/s41467-021-21784-2] [Citation(s) in RCA: 7] [Impact Index Per Article: 1.8] [Reference Citation Analysis] [Abstract] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 01/18/2021] [Accepted: 02/12/2021] [Indexed: 12/13/2022] Open
Abstract
Hyperactivation of the MAPK signaling pathway motivates the clinical use of MAPK inhibitors for BRAF-mutant melanomas. Heterogeneity in differentiation state due to epigenetic plasticity, however, results in cell-to-cell variability in the state of MAPK dependency, diminishing the efficacy of MAPK inhibitors. To identify key regulators of such variability, we screen 276 epigenetic-modifying compounds, individually or combined with MAPK inhibitors, across genetically diverse and isogenic populations of melanoma cells. Following single-cell analysis and multivariate modeling, we identify three classes of epigenetic inhibitors that target distinct epigenetic states associated with either one of the lysine-specific histone demethylases Kdm1a or Kdm4b, or BET bromodomain proteins. While melanocytes remain insensitive, the anti-tumor efficacy of each inhibitor is predicted based on melanoma cells' differentiation state and MAPK activity. Our systems pharmacology approach highlights a path toward identifying actionable epigenetic factors that extend the BRAF oncogene addiction paradigm on the basis of tumor cell differentiation state.
Collapse
Affiliation(s)
- Mehwish Khaliq
- Department of Biomedical Engineering, University of Virginia, Charlottesville, VA, USA
| | - Mohan Manikkam
- Department of Biomedical Engineering, University of Virginia, Charlottesville, VA, USA
| | - Elisabeth D Martinez
- Department of Pharmacology, UT Southwestern Medical Center, Dallas, TX, USA.,Hamon Center for Therapeutic Oncology Research, UT Southwestern Medical Center, Dallas, TX, USA
| | | |
Collapse
|
|
42
|
Oner E, Kotmakci M, Baird AM, Gray SG, Debelec Butuner B, Bozkurt E, Kantarci AG, Finn SP. Development of EphA2 siRNA-loaded lipid nanoparticles and combination with a small-molecule histone demethylase inhibitor in prostate cancer cells and tumor spheroids. J Nanobiotechnology 2021; 19:71. [PMID: 33685469 PMCID: PMC7938557 DOI: 10.1186/s12951-021-00781-z] [Citation(s) in RCA: 24] [Impact Index Per Article: 6.0] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 09/29/2020] [Accepted: 01/22/2021] [Indexed: 12/09/2022] Open
Abstract
BACKGROUND siRNAs hold a great potential for cancer therapy, however, poor stability in body fluids and low cellular uptake limit their use in the clinic. To enhance the bioavailability of siRNAs in tumors, novel, safe, and effective carriers are needed. RESULTS Here, we developed cationic solid lipid nanoparticles (cSLNs) to carry siRNAs targeting EphA2 receptor tyrosine kinase (siEphA2), which is overexpressed in many solid tumors including prostate cancer. Using DDAB cationic lipid instead of DOTMA reduced nanoparticle size and enhanced both cellular uptake and gene silencing in prostate cancer cells. DDAB-cSLN showed better cellular uptake efficiency with similar silencing compared to commercial transfection reagent (Dharmafect 2). After verifying the efficacy of siEphA2-loaded nanoparticles, we further evaluated a potential combination with a histone lysine demethylase inhibitor, JIB-04. Silencing EphA2 by siEphA2-loaded DDAB-cSLN did not affect the viability (2D or 3D culture), migration, nor clonogenicity of PC-3 cells alone. However, upon co-administration with JIB-04, there was a decrease in cellular responses. Furthermore, JIB-04 decreased EphA2 expression, and thus, silencing by siEphA2-loaded nanoparticles was further increased with co-treatment. CONCLUSIONS We have successfully developed a novel siRNA-loaded lipid nanoparticle for targeting EphA2. Moreover, preliminary results of the effects of JIB-04, alone and in combination with siEphA2, on prostate cancer cells and prostate cancer tumor spheroids were presented for the first time. Our delivery system provides high transfection efficiency and shows great promise for targeting other genes and cancer types in further in vitro and in vivo studies.
Collapse
Affiliation(s)
- Ezgi Oner
- Department of Histopathology and Morbid Anatomy, Sir Patrick Dun Translational Research Lab, St. James's Hospital, Dublin, Ireland.,Department of Pharmaceutical Biotechnology, Faculty of Pharmacy, Ege University, Bornova, Izmir, Turkey.,Department of Pharmaceutical Biotechnology, Faculty of Pharmacy, Izmir Katip Celebi University, Balatcik, Izmir, Turkey
| | - Mustafa Kotmakci
- Department of Pharmaceutical Biotechnology, Faculty of Pharmacy, Ege University, Bornova, Izmir, Turkey
| | - Anne-Marie Baird
- Department of Histopathology and Morbid Anatomy, Sir Patrick Dun Translational Research Lab, St. James's Hospital, Dublin, Ireland.,Thoracic Oncology Research Group, Trinity Translational Medicine Institute, St. James's Hospital, Dublin, Ireland.,Department of Clinical Medicine, Trinity College Dublin, Dublin, Ireland
| | - Steven G Gray
- Thoracic Oncology Research Group, Trinity Translational Medicine Institute, St. James's Hospital, Dublin, Ireland.,Department of Clinical Medicine, Trinity College Dublin, Dublin, Ireland
| | - Bilge Debelec Butuner
- Department of Pharmaceutical Biotechnology, Faculty of Pharmacy, Ege University, Bornova, Izmir, Turkey
| | - Emir Bozkurt
- Department of Genetics and Bioengineering, Faculty of Engineering, Izmir University of Economics, Balcova, Izmir, Turkey
| | - Ayse Gulten Kantarci
- Department of Pharmaceutical Biotechnology, Faculty of Pharmacy, Ege University, Bornova, Izmir, Turkey
| | - Stephen P Finn
- Department of Histopathology and Morbid Anatomy, Sir Patrick Dun Translational Research Lab, St. James's Hospital, Dublin, Ireland. .,Thoracic Oncology Research Group, Trinity Translational Medicine Institute, St. James's Hospital, Dublin, Ireland. .,Department of Histopathology, Labmed Directorate, St. James's Hospital, Dublin, Ireland. .,Cancer Molecular Diagnostics, Labmed Directorate, St. James's Hospital, Dublin, Ireland.
| |
Collapse
|
|
43
|
Macedo-Silva C, Miranda-Gonçalves V, Lameirinhas A, Lencart J, Pereira A, Lobo J, Guimarães R, Martins AT, Henrique R, Bravo I, Jerónimo C. JmjC-KDMs KDM3A and KDM6B modulate radioresistance under hypoxic conditions in esophageal squamous cell carcinoma. Cell Death Dis 2020; 11:1068. [PMID: 33318475 PMCID: PMC7736883 DOI: 10.1038/s41419-020-03279-y] [Citation(s) in RCA: 32] [Impact Index Per Article: 6.4] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 04/08/2020] [Revised: 10/30/2020] [Accepted: 11/03/2020] [Indexed: 12/24/2022]
Abstract
Esophageal squamous cell carcinoma (ESCC), the most frequent esophageal cancer (EC) subtype, entails dismal prognosis. Hypoxia, a common feature of advanced ESCC, is involved in resistance to radiotherapy (RT). RT response in hypoxia might be modulated through epigenetic mechanisms, constituting novel targets to improve patient outcome. Post-translational methylation in histone can be partially modulated by histone lysine demethylases (KDMs), which specifically removes methyl groups in certain lysine residues. KDMs deregulation was associated with tumor aggressiveness and therapy failure. Thus, we sought to unveil the role of Jumonji C domain histone lysine demethylases (JmjC-KDMs) in ESCC radioresistance acquisition. The effectiveness of RT upon ESCC cells under hypoxic conditions was assessed by colony formation assay. KDM3A/KDM6B expression, and respective H3K9me2 and H3K27me3 target marks, were evaluated by RT-qPCR, Western blot, and immunofluorescence. Effect of JmjC-KDM inhibitor IOX1, as well as KDM3A knockdown, in in vitro functional cell behavior and RT response was assessed in ESCC under hypoxic conditions. In vivo effect of combined IOX1 and ionizing radiation treatment was evaluated in ESCC cells using CAM assay. KDM3A, KDM6B, HIF-1α, and CAIX immunoexpression was assessed in primary ESCC and normal esophagus. Herein, we found that hypoxia promoted ESCC radioresistance through increased KDM3A/KDM6B expression, enhancing cell survival and migration and decreasing DNA damage and apoptosis, in vitro. Exposure to IOX1 reverted these features, increasing ESCC radiosensitivity and decreasing ESCC microtumors size, in vivo. KDM3A was upregulated in ESCC tissues compared to the normal esophagus, associating and colocalizing with hypoxic markers (HIF-1α and CAIX). Therefore, KDM3A upregulation in ESCC cell lines and primary tumors associated with hypoxia, playing a critical role in EC aggressiveness and radioresistance. KDM3A targeting, concomitant with conventional RT, constitutes a promising strategy to improve ESCC patients' survival.
Collapse
Affiliation(s)
- Catarina Macedo-Silva
- Cancer Biology & Epigenetics Group - Research Center, Portuguese Oncology Institute of Porto (CI-IPOP), Porto, Portugal
| | - Vera Miranda-Gonçalves
- Cancer Biology & Epigenetics Group - Research Center, Portuguese Oncology Institute of Porto (CI-IPOP), Porto, Portugal
| | - Ana Lameirinhas
- Cancer Biology & Epigenetics Group - Research Center, Portuguese Oncology Institute of Porto (CI-IPOP), Porto, Portugal
| | - Joana Lencart
- Medical Physics, Radiobiology and Radiation Protection Group - Research Center, Portuguese Oncology Institute of Porto (CI-IPOP), Porto, Portugal
- Departments of Medical Physics, Portuguese Oncology Institute of Porto, Porto, Portugal
| | - Alexandre Pereira
- Medical Physics, Radiobiology and Radiation Protection Group - Research Center, Portuguese Oncology Institute of Porto (CI-IPOP), Porto, Portugal
- Departments of Medical Physics, Portuguese Oncology Institute of Porto, Porto, Portugal
| | - João Lobo
- Cancer Biology & Epigenetics Group - Research Center, Portuguese Oncology Institute of Porto (CI-IPOP), Porto, Portugal
- Departments of Pathology, Portuguese Oncology Institute of Porto, Porto, Portugal
- Department of Pathology and Molecular Immunology, Institute of Biomedical Sciences Abel Salazar - University of Porto (ICBAS-UP), Porto, Portugal
| | - Rita Guimarães
- Departments of Pathology, Portuguese Oncology Institute of Porto, Porto, Portugal
| | - Ana Teresa Martins
- Departments of Pathology, Portuguese Oncology Institute of Porto, Porto, Portugal
| | - Rui Henrique
- Cancer Biology & Epigenetics Group - Research Center, Portuguese Oncology Institute of Porto (CI-IPOP), Porto, Portugal
- Departments of Pathology, Portuguese Oncology Institute of Porto, Porto, Portugal
- Department of Pathology and Molecular Immunology, Institute of Biomedical Sciences Abel Salazar - University of Porto (ICBAS-UP), Porto, Portugal
| | - Isabel Bravo
- Medical Physics, Radiobiology and Radiation Protection Group - Research Center, Portuguese Oncology Institute of Porto (CI-IPOP), Porto, Portugal
| | - Carmen Jerónimo
- Cancer Biology & Epigenetics Group - Research Center, Portuguese Oncology Institute of Porto (CI-IPOP), Porto, Portugal.
- Department of Pathology and Molecular Immunology, Institute of Biomedical Sciences Abel Salazar - University of Porto (ICBAS-UP), Porto, Portugal.
| |
Collapse
|
|
44
|
Zhou Y, Shao C. Histone methylation can either promote or reduce cellular radiosensitivity by regulating DNA repair pathways. MUTATION RESEARCH-REVIEWS IN MUTATION RESEARCH 2020; 787:108362. [PMID: 34083050 DOI: 10.1016/j.mrrev.2020.108362] [Citation(s) in RCA: 4] [Impact Index Per Article: 0.8] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Subscribe] [Scholar Register] [Received: 07/27/2020] [Revised: 12/03/2020] [Accepted: 12/07/2020] [Indexed: 10/22/2022]
Abstract
Radiotherapy is one of the primary modalities for cancer treatment, and its efficiency usually relies on cellular radiosensitivity. DNA damage repair is a core content of cellular radiosensitivity, and the primary mechanism of which includes non-homologous end-joining (NHEJ) and homologous recombination (HR). By affecting DNA damage repair, histone methylation regulated by histone methyltransferases (HMTs) and histone demethylases (HDMs) participates in the regulation of cellular radiosensitivity via three mechanisms: (a) recruiting DNA repair-related proteins, (b) regulating the expressions of DNA repair genes, and (c) mediating the dynamic change of chromatin. Interestingly, both aberrantly high and low levels of histone methylation could impede DNA repair processes. Here we reviewed the mechanisms of the dual effects of histone methylation on cell response to radiation. Since some inhibitors of HMTs and HDMs are reported to increase cellular radiosensitivity, understanding their molecular mechanisms may be helpful in developing new drugs for the therapy of radioresistant tumors.
Collapse
Affiliation(s)
- Yuchuan Zhou
- Institute of Radiation Medicine, Shanghai Medical College, Fudan University, No. 2094 Xie-Tu Road, Shanghai, 200032, China
| | - Chunlin Shao
- Institute of Radiation Medicine, Shanghai Medical College, Fudan University, No. 2094 Xie-Tu Road, Shanghai, 200032, China.
| |
Collapse
|
|
45
|
Xu LM, Yu H, Yuan YJ, Zhang J, Ma Y, Cao XC, Wang J, Zhao LJ, Wang P. Overcoming of Radioresistance in Non-small Cell Lung Cancer by microRNA-320a Through HIF1α-Suppression Mediated Methylation of PTEN. Front Cell Dev Biol 2020; 8:553733. [PMID: 33304897 PMCID: PMC7693713 DOI: 10.3389/fcell.2020.553733] [Citation(s) in RCA: 17] [Impact Index Per Article: 3.4] [Reference Citation Analysis] [Abstract] [Key Words] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 04/20/2020] [Accepted: 08/18/2020] [Indexed: 11/13/2022] Open
Abstract
Background Radioresistance is a major challenge in the use of radiotherapy for the treatment of lung cancer while microRNAs (miRs) have been reported to participate in multiple essential cellular processes including radiosensitization. This study was conducted with the main objective of investigating the potential role of miR-320a in radioresistance of non-small cell lung cancer (NSCLC) via the possible mechanism related to HIF1α, KDM5B, and PTEN. Methods Firstly, NSCLC radiosensitivity-related microarray dataset GSE112374 was obtained. Then, the expression of miR-320a, HIF1α, KDM5B, and PTEN was detected in the collected clinical NSCLC samples, followed by Pearson's correlation analysis. Subsequently, ChIP assay was conducted to determine the content of the PTEN promoter fragment enriched by the IgG antibody and H3K4me3 antibody. Finally, a series of in vitro and in vivo assays were performed in order to evaluate the effects of miR-320a on radioresistance of NSCLC with the involvement of HIF1α, KDM5B, and PTEN. Results The microarray dataset GSE112374 presented with a high expression of miR-320a in NSCLC radiosensitivity samples, which was further confirmed in our clinical samples with the use of reverse transcription-quantitative polymerase chain reaction. Moreover, miR-320a negatively targeted HIF1α, inhibiting radioresistance of NSCLC. Interestingly, miR-320a suppressed the expression of KDM5B, and KDM5B was found to enhance the radioresistance of NSCLC through the downregulation of PTEN expression. The inhibition of miR-320a in radioresistance of NSCLC was also reproduced by in vivo assay. Conclusion Taken together, our findings were suggestive of the inhibitory effect of miR-320a on radioresistance of NSCLC through HIF1α-suppression mediated methylation of PTEN.
Collapse
Affiliation(s)
- Li-Ming Xu
- Department of Radiotherapy, Tianjin Medical University Cancer Institute & Hospital, National Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Breast Cancer Prevention and Therapy, Tianjin Medical University, Ministry of Education, Tianjin, China.,Department of Radiotherapy, Tianjin Medical University Cancer Hospital Airport Hospital, Tianjin, China
| | - Hao Yu
- Department of Radiotherapy, Tianjin Medical University Cancer Institute & Hospital, National Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Breast Cancer Prevention and Therapy, Tianjin Medical University, Ministry of Education, Tianjin, China
| | - Ya-Jing Yuan
- Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, Tianjin, China.,Department of Anesthesia, Tianjin Medical University Cancer Institute & Hospital, National Clinical Research Center for Cancer, Tianjin, China
| | - Jiao Zhang
- Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Breast Cancer Prevention and Therapy, Tianjin Medical University, Ministry of Education, Tianjin, China.,The First Department of Breast Cancer, Tianjin Medical University Cancer Institute & Hospital, National Clinical Research Center for Cancer, Tianjin, China
| | - Yue Ma
- Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Breast Cancer Prevention and Therapy, Tianjin Medical University, Ministry of Education, Tianjin, China.,The First Department of Breast Cancer, Tianjin Medical University Cancer Institute & Hospital, National Clinical Research Center for Cancer, Tianjin, China
| | - Xu-Chen Cao
- Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Breast Cancer Prevention and Therapy, Tianjin Medical University, Ministry of Education, Tianjin, China.,The First Department of Breast Cancer, Tianjin Medical University Cancer Institute & Hospital, National Clinical Research Center for Cancer, Tianjin, China
| | - Jun Wang
- Department of Radiotherapy, Tianjin Medical University Cancer Institute & Hospital, National Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Breast Cancer Prevention and Therapy, Tianjin Medical University, Ministry of Education, Tianjin, China.,Department of Radiotherapy, Tianjin Medical University Cancer Hospital Airport Hospital, Tianjin, China
| | - Lu-Jun Zhao
- Department of Radiotherapy, Tianjin Medical University Cancer Institute & Hospital, National Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Breast Cancer Prevention and Therapy, Tianjin Medical University, Ministry of Education, Tianjin, China
| | - Ping Wang
- Department of Radiotherapy, Tianjin Medical University Cancer Institute & Hospital, National Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, Tianjin, China.,Key Laboratory of Breast Cancer Prevention and Therapy, Tianjin Medical University, Ministry of Education, Tianjin, China
| |
Collapse
|
|
46
|
Wang Y, Ma J, Martinez ED, Liang D, Xie H. A UHPLC-MS/MS method for the quantification of JIB-04 in rat plasma: Development, validation and application to pharmacokinetics study. J Pharm Biomed Anal 2020; 191:113587. [PMID: 32892084 PMCID: PMC7581536 DOI: 10.1016/j.jpba.2020.113587] [Citation(s) in RCA: 3] [Impact Index Per Article: 0.6] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Journal Information] [Subscribe] [Scholar Register] [Received: 06/26/2020] [Revised: 08/18/2020] [Accepted: 08/20/2020] [Indexed: 12/14/2022]
Abstract
Methylation of lysine by histone methyltransferases can be reversed by lysine demethylases (KDMs). Different KDMs have distinct oncogenic functions based on their cellular localization, stimulating cancer cell proliferation, reducing the expression of tumor suppressors, and/or promoting the development of drug resistance. JIB-04 is a small molecule that pan-selectively inhibits KDMs, showing maximal inhibitory activity against KDM5A, and as secondary targets, KDM4D/4B/4A/6B/4C. Recently, it was found that JIB-04 also potently and selectively blocks HIV-1 Tat expression, transactivation, and virus replication in T cell lines via the inhibition of a new target, serine hydroxymethyltransferase 2. Pharmacokinetic characterization and an analytical method for the quantification of JIB-04 are necessary for the further development of this small molecule. Herein, a sensitive, specific, fast and reliable UHPLC-MS/MS method for the quantification of JIB-04 in rat plasma samples was developed and fully validated using a SCIEX 6500+ triple QUAD LC-MS system equipped with an ExionLC UHPLC unit. The chromatographic separation was achieved on a reverse phase ACE Excel 2 Super C18 column with a flow rate of 0.5 mL/min under gradient elution. The calibration curves were linear (r2 > 0.999) over concentrations from 0.5 to 1000 ng/mL. The accuracy (RE%) was between -7.4% and 3.7%, and the precision (CV%) was 10.2% or less. The stability data showed that no significant degradation occurred under the experimental conditions. This method was successfully applied to the pharmacokinetic study of JIB-04 in rat plasma after intravenous and oral administration and the oral bioavailability of JIB-04 was found to be 44.4%.
Collapse
Affiliation(s)
- Yang Wang
- Department of Pharmaceutical and Environmental Health Sciences, College of Pharmacy and Health Sciences, Texas Southern University, Houston, TX 77004, USA
| | - Jing Ma
- Department of Pharmaceutical and Environmental Health Sciences, College of Pharmacy and Health Sciences, Texas Southern University, Houston, TX 77004, USA
| | - Elisabeth D Martinez
- Department of Pharmacology, UT Southwestern Medical Center, Dallas, TX 75390, USA
| | - Dong Liang
- Department of Pharmaceutical and Environmental Health Sciences, College of Pharmacy and Health Sciences, Texas Southern University, Houston, TX 77004, USA
| | - Huan Xie
- Department of Pharmaceutical and Environmental Health Sciences, College of Pharmacy and Health Sciences, Texas Southern University, Houston, TX 77004, USA.
| |
Collapse
|
|
47
|
ASH2L drives proliferation and sensitivity to bleomycin and other genotoxins in Hodgkin's lymphoma and testicular cancer cells. Cell Death Dis 2020; 11:1019. [PMID: 33257682 PMCID: PMC7705021 DOI: 10.1038/s41419-020-03231-0] [Citation(s) in RCA: 5] [Impact Index Per Article: 1.0] [Reference Citation Analysis] [Abstract] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 08/20/2020] [Revised: 11/12/2020] [Accepted: 11/12/2020] [Indexed: 12/24/2022]
Abstract
It is of clinical importance to identify biomarkers predicting the efficacy of DNA damaging drugs (genotoxins) so that nonresponders are not unduly exposed to the deleterious effects of otherwise inefficient drugs. Here, we initially focused on the bleomycin genotoxin because of the limited information about the genes implicated in the sensitivity or resistance to this compound. Using a whole-genome CRISPR/Cas9 gene knockout approach, we identified ASH2L, a core component of the H3K4 methyl transferase complex, as a protein required for bleomycin sensitivity in L1236 Hodgkin lymphoma. Knocking down ASH2L in these cells and in the NT2D1 testicular cancer cell line rendered them resistant to bleomycin, etoposide, and cisplatin but did not affect their sensitivity toward ATM or ATR inhibitors. ASH2L knockdown decreased cell proliferation and facilitated DNA repair via homologous recombination and nonhomologous end-joining mechanisms. Data from the Tumor Cancer Genome Atlas indicate that patients with testicular cancer carrying alterations in the ASH2L gene are more likely to relapse than patients with unaltered ASH2L genes. The cell models we have used are derived from cancers currently treated either partially (Hodgkin’s lymphoma), or entirely (testicular cancer) with genotoxins. For such cancers, ASH2L levels could be used as a biomarker to predict the response to genotoxins. In situations where tumors are expressing low levels of ASH2L, which may allow them to resist genotoxic treatment, the use of ATR or ATM inhibitors may be more efficacious as our data indicate that ASH2L knockdown does not affect sensitivity to these inhibitors.
Collapse
|
|
48
|
Liu B, Kumar R, Chao HP, Mehmood R, Ji Y, Tracz A, Tang DG. Evidence for context-dependent functions of KDM5B in prostate development and prostate cancer. Oncotarget 2020; 11:4243-4252. [PMID: 33245716 PMCID: PMC7679033 DOI: 10.18632/oncotarget.27818] [Citation(s) in RCA: 8] [Impact Index Per Article: 1.6] [Reference Citation Analysis] [Abstract] [Key Words] [Grants] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 08/20/2020] [Accepted: 10/29/2020] [Indexed: 01/09/2023] Open
Abstract
Prostate cancer (PCa) is one of the leading causes of cancer-related deaths worldwide. Prostate tumorigenesis and PCa progression involve numerous genetic as well as epigenetic perturbations. Histone modification represents a fundamental epigenetic mechanism that regulates diverse cellular processes, and H3K4 methylation, one such histone modification associated with active transcription, can be reversed by dedicated histone demethylase KDM5B (JARID1B). Abnormal expression and functions of KDM5B have been implicated in several cancer types including PCa. Consistently, our bioinformatics analysis reveals that the KDM5B mRNA levels are upregulated in PCa compared to benign prostate tissues, and correlate with increased tumor grade and poor patient survival, supporting an oncogenic function of KDM5B in PCa. Surprisingly, however, when we generated prostate-specific conditional Kdm5b knockout mice using probasin (Pb) promoter-driven Cre: loxP system, we observed that Kdm5b deletion did not affect normal prostate development but instead induced mild hyperplasia. These results suggest that KDM5B may possess context-dependent roles in normal prostate development vs. PCa development and progression.
Collapse
Affiliation(s)
- Bigang Liu
- Department of Epigenetics and Molecular Carcinogenesis, The University of Texas M.D Anderson Cancer Center, Science Park, Smithville, TX, USA
- These authors contributed equally to this work
| | - Rahul Kumar
- Department of Pharmacology & Therapeutics, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA
- These authors contributed equally to this work
| | - Hseuh-Ping Chao
- Department of Epigenetics and Molecular Carcinogenesis, The University of Texas M.D Anderson Cancer Center, Science Park, Smithville, TX, USA
- Livestrong Cancer Institutes, Dell Medical School, The University of Texas at Austin, Austin, TX, USA
| | - Rashid Mehmood
- Department of Pharmacology & Therapeutics, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA
- Department of Life Sciences, College of Science and General Studies, Alfaisal University, Takhasusi Street, Riyadh, Saudi Arabia
| | - Yibing Ji
- Department of Epigenetics and Molecular Carcinogenesis, The University of Texas M.D Anderson Cancer Center, Science Park, Smithville, TX, USA
| | - Amanda Tracz
- Department of Epigenetics and Molecular Carcinogenesis, The University of Texas M.D Anderson Cancer Center, Science Park, Smithville, TX, USA
| | - Dean G. Tang
- Department of Epigenetics and Molecular Carcinogenesis, The University of Texas M.D Anderson Cancer Center, Science Park, Smithville, TX, USA
- Department of Pharmacology & Therapeutics, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA
| |
Collapse
|
|
49
|
Götting I, Jendrossek V, Matschke J. A New Twist in Protein Kinase B/Akt Signaling: Role of Altered Cancer Cell Metabolism in Akt-Mediated Therapy Resistance. Int J Mol Sci 2020; 21:ijms21228563. [PMID: 33202866 PMCID: PMC7697684 DOI: 10.3390/ijms21228563] [Citation(s) in RCA: 18] [Impact Index Per Article: 3.6] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 09/16/2020] [Revised: 10/23/2020] [Accepted: 11/09/2020] [Indexed: 12/11/2022] Open
Abstract
Cancer resistance to chemotherapy, radiotherapy and molecular-targeted agents is a major obstacle to successful cancer therapy. Herein, aberrant activation of the phosphatidyl-inositol-3-kinase (PI3K)/protein kinase B (Akt) pathway is one of the most frequently deregulated pathways in cancer cells and has been associated with multiple aspects of therapy resistance. These include, for example, survival under stress conditions, apoptosis resistance, activation of the cellular response to DNA damage and repair of radiation-induced or chemotherapy-induced DNA damage, particularly DNA double strand breaks (DSB). One further important, yet not much investigated aspect of Akt-dependent signaling is the regulation of cell metabolism. In fact, many Akt target proteins are part of or involved in the regulation of metabolic pathways. Furthermore, recent studies revealed the importance of certain metabolites for protection against therapy-induced cell stress and the repair of therapy-induced DNA damage. Thus far, the likely interaction between deregulated activation of Akt, altered cancer metabolism and therapy resistance is not yet well understood. The present review describes the documented interactions between Akt, its target proteins and cancer cell metabolism, focusing on antioxidant defense and DSB repair. Furthermore, the review highlights potential connections between deregulated Akt, cancer cell metabolism and therapy resistance of cancer cells through altered DSB repair and discusses potential resulting therapeutic implications.
Collapse
|
|
50
|
Xiang K, Jendrossek V, Matschke J. Oncometabolites and the response to radiotherapy. Radiat Oncol 2020; 15:197. [PMID: 32799884 PMCID: PMC7429799 DOI: 10.1186/s13014-020-01638-9] [Citation(s) in RCA: 15] [Impact Index Per Article: 3.0] [Reference Citation Analysis] [Abstract] [Key Words] [MESH Headings] [Grants] [Track Full Text] [Download PDF] [Figures] [Journal Information] [Subscribe] [Scholar Register] [Received: 05/15/2020] [Accepted: 08/06/2020] [Indexed: 12/18/2022] Open
Abstract
Radiotherapy (RT) is applied in 45-60% of all cancer patients either alone or in multimodal therapy concepts comprising surgery, RT and chemotherapy. However, despite technical innovations approximately only 50% are cured, highlight a high medical need for innovation in RT practice. RT is a multidisciplinary treatment involving medicine and physics, but has always been successful in integrating emerging novel concepts from cancer and radiation biology for improving therapy outcome. Currently, substantial improvements are expected from integration of precision medicine approaches into RT concepts.Altered metabolism is an important feature of cancer cells and a driving force for malignant progression. Proper metabolic processes are essential to maintain and drive all energy-demanding cellular processes, e.g. repair of DNA double-strand breaks (DSBs). Consequently, metabolic bottlenecks might allow therapeutic intervention in cancer patients.Increasing evidence now indicates that oncogenic activation of metabolic enzymes, oncogenic activities of mutated metabolic enzymes, or adverse conditions in the tumor microenvironment can result in abnormal production of metabolites promoting cancer progression, e.g. 2-hyroxyglutarate (2-HG), succinate and fumarate, respectively. Interestingly, these so-called "oncometabolites" not only modulate cell signaling but also impact the response of cancer cells to chemotherapy and RT, presumably by epigenetic modulation of DNA repair.Here we aimed to introduce the biological basis of oncometabolite production and of their actions on epigenetic regulation of DNA repair. Furthermore, the review will highlight innovative therapeutic opportunities arising from the interaction of oncometabolites with DNA repair regulation for specifically enhancing the therapeutic effects of genotoxic treatments including RT in cancer patients.
Collapse
Affiliation(s)
- Kexu Xiang
- Institute of Cell Biology (Cancer Research), University Hospital Essen, University of Duisburg-Essen, Virchowstrasse 173, 45147, Essen, Germany
| | - Verena Jendrossek
- Institute of Cell Biology (Cancer Research), University Hospital Essen, University of Duisburg-Essen, Virchowstrasse 173, 45147, Essen, Germany
| | - Johann Matschke
- Institute of Cell Biology (Cancer Research), University Hospital Essen, University of Duisburg-Essen, Virchowstrasse 173, 45147, Essen, Germany.
| |
Collapse
|